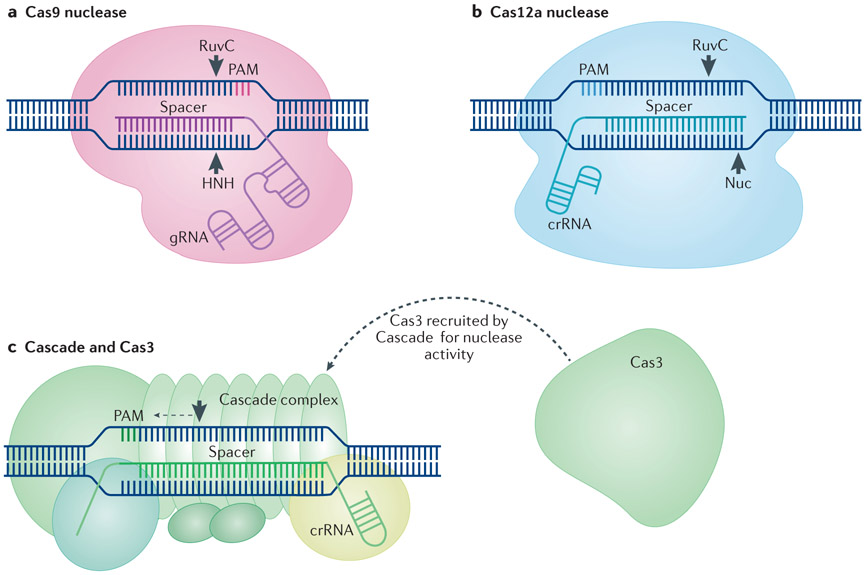
Figure 1. Overview of the main CRISPR–Cas gene editing tools.
a ∣ CRISPR-associated endonuclease 9 (Cas9) proteins rely on RNA guidance for targeting specificity. In engineered CRISPR–Cas9 systems, Cas9 interacts with the backbone of the guide RNA (gRNA). Complementary pairing of the spacer portion of the gRNA to a DNA target sequence positioned next to a 5’ protospacer adjacent motif (PAM) results in generation of a blunt DNA double-strand break by the two Cas9 nuclease domains, RuvC and HNH11-13. b ∣ Cas12a nucleases recognize DNA target sequences with complementarity to the crRNA spacer positioned next to a 3’ PAM. Target recognition results in generation of a staggered DNA double-strand break by a RuvC domain and a putative nuclease (Nuc) domain33. c ∣ Cascade is a multimeric complex that targets DNA that has complementarity to the spacer portion of a crRNA and that is positioned next to a 3’ PAM37-42. Following target recognition, Cascade recruits Cas3 to generate a single-strand nick, which is followed by 3’ to 5’ degradation of the targeted DNA37,39,44,45.