Figure 4. EBF2 Cooperates with ERRα and PGC1α to Promote Ucp1 Transcription.
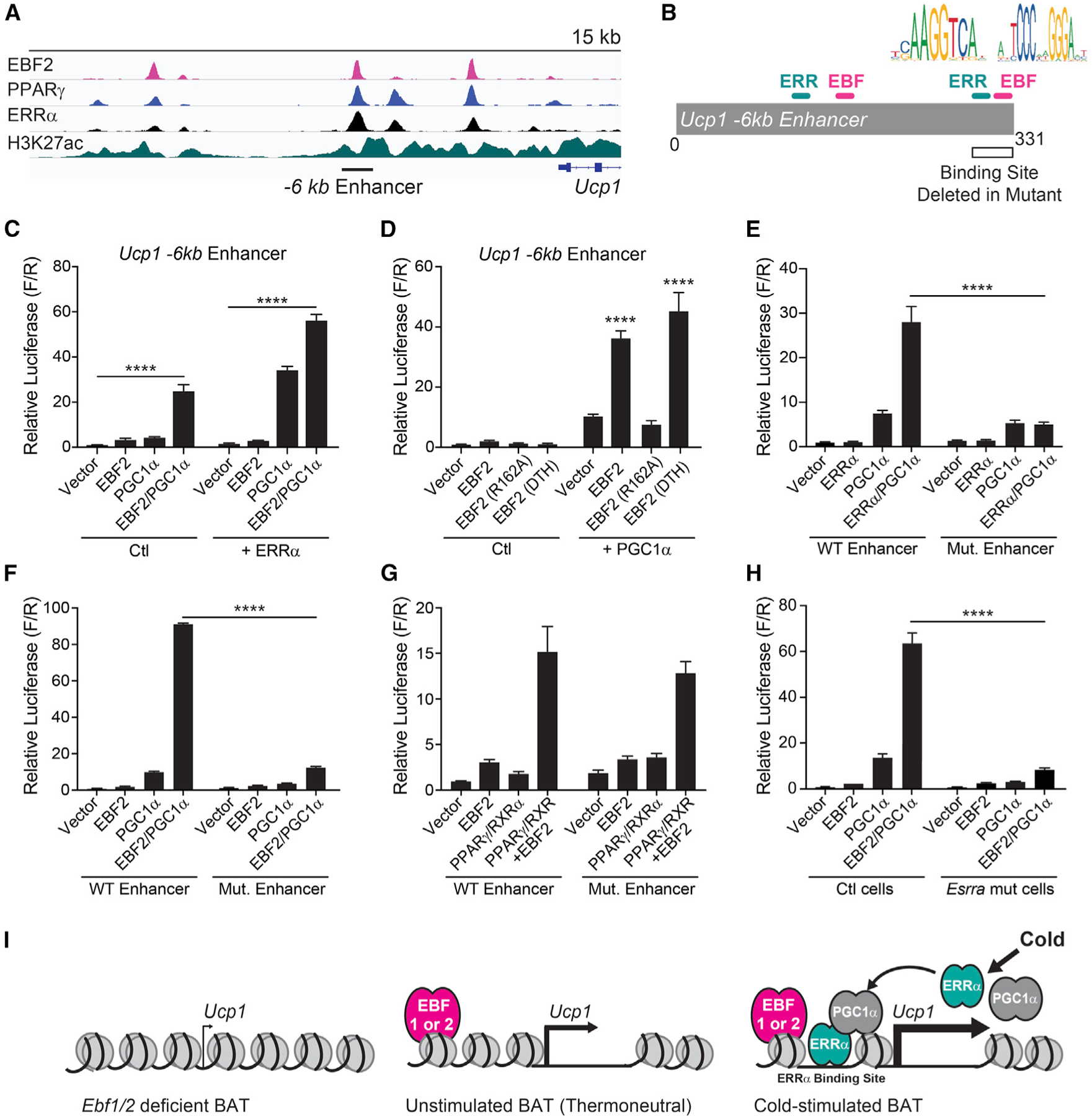
(A) Chromatin immunoprecipitation sequencing (ChIP-seq) tracks of indicated transcription factors and histone modifications at the Ucp1 locus (Emmett et al., 2017; Shapira et al., 2017).
(B) Schematic depicting Ucp1 −6 kb reporter construct and putative EBF and ERR binding sites deleted in mutant reporter.
(C and D) Transcriptional activity of the Ucp1 −6 kb enhancer in NIH 3T3 cells upon expression of (C) EBF2, PGC1α, and/or ERRα; (D) EBF2, EBF2 R162A (DNA-binding mutant) or EBF2 DTH (C-terminal domain mutant); and/or PGC1α (n = 3 replicates per group; mean ± SEM).
(E–G) Transcriptional activity of the WT or mutant (Mut.) Ucp1 −6 kb enhancer in NIH 3T3 cells upon expression of (E) ERRα and/or PGC1α (n = 3); (F) EBF2 and/or PGC1α (n = 3); or (G) EBF2 and/or PPARγ/RXRα (n = 9) (mean ± SEM).
(H) Transcriptional activity of the Ucp1 −6 kb enhancer in control (ctl) or Esrra-deficient NIH 3T3 cells upon expression of EBF2 and/or PGC1α (n = 3 replicates per group; mean ± SEM).
(I) Model of EBF function at thermogenic genes (i.e., Ucp1) in the context of cold exposure.
