Abstract
Swine enteric coronaviruses are a group of most significant pathogens causing diarrhea in piglets with similar clinical symptoms and pathological changes. To develop a simple, rapid, accurate, and high-throughput detection method for diagnosis and differential diagnosis on swine enteric coronaviruses, specific primers and probes were designed based on the highly conserved regions of transmissible gastroenteritis virus (TGEV) N, porcine epidemic diarrhea virus (PEDV) M, porcine deltacoronavirus (PDCoV) M, and porcine enteric alphacoronavirus (PEAV) N genes respectively. A TaqMan-probe-based multiplex real-time RT-qPCR assay was developed and optimized to simultaneously detect these swine enteric coronaviruses. The results showed that the limit of detection can reach as low as 10 copies in singular real-time RT-qPCR assays and 100 copies in multiplex real-time RT-qPCR assay, with all correlation coefficients (R2) at above 0.99, and the amplification efficiency at between 90 and 120%. This multiplex real-time RT-qPCR assay demonstrated high sensitivity, extreme specificity, and excellent repeatability. The multiplex real-time RT-qPCR assay was then employed to detect the swine enteric coronavirus from 354 field diarrheal samples. The results manifested that TGEV and PDCoV were the main pathogens in these samples, accompanied by co-infections. This well-established multiplex real-time RT-qPCR assay provided a rapid, efficient, specific, and sensitive tool for detection of swine enteric coronaviruses.
Electronic supplementary material
The online version of this article (10.1007/s00253-019-09835-7) contains supplementary material, which is available to authorized users.
Keywords: TaqMan probe, Multiplex real-time RT-qPCR, Diagnosis, Swine enteric coronaviruses
Introduction
Porcine enteric viruses, the pathogens of viral diarrhea in pigs, caused huge economic losses in the swine industry in recent years (Zhang et al. 2016). There were more than ten enteric viruses discovered from swine gut in the past, including but not limited to transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), porcine deltacoronavirus (PDCoV), porcine enteric alphacoronavirus (PEAV), porcine sapovirus (PSaV), porcine norovirus (PNoV), porcine teschenvirus (PTV), porcine kobuvirus (PKV), Seneca Valley virus (SVV), porcine rotavirus (PRV), porcine reovirus (ReoV), porcine bocavirus (PBoV), porcine astrovirus (PAstV), and etc.
Within these swine enteric viruses, coronaviruses are the most devastating pathogens responsible for acute diarrhea, vomiting, dehydration, and high mortality in neonatal and suckling piglets. According to the genetic and antigenic characters, coronavirus was divided into four genera: Alpha-, Beta-, Gamma-, and Delta-CoV (Woo et al. 2010). During the past years, some alphacoronaviruses (PEDV, TGEV, and PEAV) and a deltacoronavirus (PDCoV) emerged or reemerged in the pig farms and resulted in severe diarrhea and mortality in the early stage of suckling piglets.
The TGEV and PEDV were the traditional causal agents responsible for diarrhea in pigs for the past decades. However, the variant strains of PEDV were discovered since 2010 and circulating in pig herds thereafter, showing up to 100% death rate for piglets younger than 1-week-old (Lyoo et al. 2017; Sun et al. 2012). Another swine enteric virus, porcine deltacoronavirus (PDCoV), was firstly discovered from healthy pig herds by a research team of Hong Kong in 2012 when they did a molecular epidemiology study in 3137 mammals and 3519 birds (Woo et al. 2012). Two years later, this virus was reported to cause severe diarrhea and/or vomiting and atrophic enteritis in the USA (Wang et al. 2014) and later on in China (Song et al. 2015). Subsequently, a novel porcine enteric alphacoronavirus PEAV was discovered in 2017 from some pig farms located in Southern China, causing more than 24,000 piglets’ death (Gong et al. 2017). This virus was also named as SeACoV (Pan et al. 2017) or SADS-CoV (Zhou et al. 2018b) by the other research groups.
All these four swine enteric coronaviruses, causing similar clinical symptoms and pathological changes in piglets, circulate in the pig herds and result in huge economic losses across the world in recent years. To develop a simple, rapid, accurate, and high-throughput detection method for diagnosis and differential diagnosis on swine enteric coronaviruses, a TaqMan-probe-based multiplex real-time RT-qPCR assay was established to simultaneously detect TGEV, PEDV, PEAV, and PDCoV from the same reaction vial. To our knowledge, this is the first multiplex real-time RT-qPCR method for swine enteric coronaviruses detection.
Materials and methods
Viruses, clinical samples, primers, and probes
The TGEV was previously propagated and preserved in our laboratory. The PEDV and PDCoV were isolated from clinical samples and confirmed by conventional PCR and DNA sequencing (GENEWIZ, Suzhou, China). The nucleoprotein gene of PEAV (GenBank access number: MF370205) was synthesized from GENEWIZ biotech company (Suzhou, China).
Field samples were collected from diarrheal piglets between 2015 and 2018 from Liaoning, Shandong, Chongqing, Shaanxi, Ningxia, and Gansu provinces. All samples were stored at − 80 °C until use.
The primers and TaqMan probes for real-time qPCR assay were designed by the software Beacon Designer 7 (PREMIER Biosoft International, Palo Alto, CA, USA). The detailed information of primers and probes were listed in Table 1.
Table 1.
Primers and probes designed for real-time RT-qPCR
| Virus | Primer/probe | Sequence (5′-3′) | Target gene | Size (bp) |
|---|---|---|---|---|
| PEDV | Forward | GATACTTTGGCCTCTTGTGT | M | 150 |
| Reverse | CACAACCGAATGCTATTGACG | |||
| Probe | FAM-TTCAGCATCCTTATGGCTTGCATC-TAMRA | |||
| PDCoV | Forward | ATTTGGACCGCAGTTGACA | M | 92 |
| Reverse | GCCCAGGATATAAAGGTCAG | |||
| Probe | Cy5-TAAGAAGGACGCAGTTTTCATTGTG-BHQ2 | |||
| TGEV | Forward | TGCCATGAACAAACCAAC | N | 81 |
| Reverse | GGCACTTTACCATCGAAT | |||
| Probe | HEX-TAGCACCACGACTACCAAGC-BHQ1 | |||
| PEAV | Forward | TCTCGGCTTACTCTAAACCC | N | 150 |
| Reverse | CATCCACCATCTCAACCTC | |||
| Probe | TexasRed-AAGACCTAAATGCTGATGCCCCA-BHQ2 |
RNA extraction and reverse transcription
All clinical samples were resuspended with phosphate-buffer saline (PBS), vortexed and centrifuged at 12,000×g at 4 °C for 10 min. An aliquot of 250 μL supernatant was applied for total RNA extraction with RNAiso reagent (Takara, Dalian, China) following the manufacturer’s instruction. The total RNA in RNase-free water was reversely transcribed into cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, USA). The cDNA was subjected to real-time qPCR analysis with the multiplex RT-qPCR method established in this study.
Construction of recombinant plasmids for standard curves
The M genes of PEDV and PDCoV, and N genes of TGEV and PEAV were constructed into pET-30a(+) vector. The recombinant plasmids were linearized by SmaI enzyme digestion and recovered by PCR purification kit. The purified recombinant plasmids were quantified by spectrophotometric analysis. The copy number of recombination plasmids was calculated by using the following formula (Huang et al. 2009):
To establish the standard curves for single coronavirus, each plasmid was diluted in a tenfold series, from 107 copies/μL to 101 copies/μL. For multiplex standard curves, each of the four linearization plasmids was adjusted to 4 × 109 copies/μL and pooled with equal volume to made 1 × 109 copies/μL of each plasmid. The pooled plasmid was then diluted serially by tenfold to establish multiplex standard curves.
Single and multiplex real-time qPCR condition
All real-time qPCR reaction systems were set to a volume of 20 μL. For single qPCR amplifying TGEV, PEDV, and PDCoV, 10 μL 2 × TransStart Probe qPCR SuperMix (TransGene, Beijing, China), 200 nM primers and probe each, 1 μL plasmid DNA template, and 6.8 μL nuclease-free water were pooled and mixed. For PEAV amplification, 10 μL 2 × TransStart Probe qPCR SuperMix (TransGene, Beijing, China), 500 nM each primer, 100 nM probe, 1 μL plasmid DNA template, and 6.8 μL nuclease-free water were pooled and mixed. All reactions were amplified on a Bio-Rad CFX96™ Real-time System (Bio-Rad, Hercules, CA, USA) at 94 °C for 30s, followed by 40 cycles of 94 °C for 5 s and 60 °C for 30s.
For reaction system of multiplex real-time PCR, 10 μL 2 × TransStart Probe qPCR SuperMix combined with all primers, probes, templates, and nuclease-free water to a final volume of 20 μL. The concentrations of each primer and probe of PEDV, PDCoV, TGEV, and PEAV were optimized for better outputs. The amplifying cycles of multiplex qPCR were carried out as same as singular real-time qPCR. The Cq value higher than 35 was considered negative. All qPCR results were analyzed by CFX Manager™ software.
Sensitivity, specificity, and repeatability analysis of multiplex RT-qPCR assay
To analyze the sensitivity of established multiplex RT-qPCR, the linearization standard plasmids prepared above were diluted tenfold serially to a final concentration between 1.1 × 107 copies/μL and 1.1 × 101 copies/μL in nuclease-free water. The diluted standard plasmids were used as templates for real-time qPCR amplification.
To estimate the specificity of this established multiplex RT-qPCR, standard DNAs, or cDNAs of major swine viruses, including porcine astrovirus (PAstV), porcine kobuvirus (PKV), type O foot-and-mouth disease virus (FMDV-O), type A foot-and-mouth disease virus (FMDV-A), porcine reproductive and respiratory syndrome virus (PRRSV), classical swine fever virus (CSFV), porcine rotavirus (PRV), porcine circovirus 2(PCV2), and pseudorabies virus (PrV) were used as templates for amplification. The nuclease-free water was served as negative template control.
To evaluate its repeatability, tenfold serially diluted standard template between 1.1 × 107 copies/μL to 1.1 × 101 copies/μL were used to test the coefficients of variation of real-time PCR. For intra-assay repeatability, all samples were triplicated. For inter-assay repeatability, the assays were repeated three times individually at different locations.
Clinical sample detection
A total of 354 fecal samples were collected from diarrheal pig farms located in Liaoning, Shandong, Chongqing, Shaanxi, Ningxia, and Gansu provinces of China between 2015 and 2018. All samples were diluted fivefold with sterile phosphate-buffered saline (PBS), vortexed, and centrifuged at 1847×g at 4 °C for 20 min. The supernatant was collected and used to extract viral RNA with Trizol reagent (Invitrogen, Carlsbad, CA, USA). The cDNAs were generated by Reverse Transcript System (Promega, Madison, WI, USA) using extracted total RNA as templates and hexamer random primers. All cDNA from clinical samples were measured by the multiplex RT-qPCR assay developed in this study.
Results
Single real-time RT-qPCR assay for individual virus
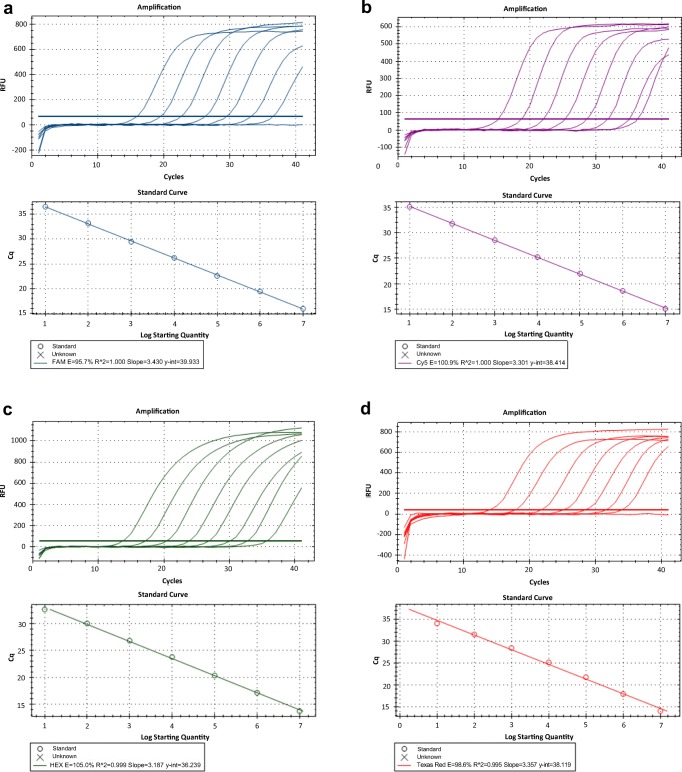
To develop a multiplex real-time RT-qPCR, the single real-time RT-qPCRs for detection of the individual virus were firstly established with different fluorescence-labeled target probes, for details, FAM for PEDV M gene, Cy5 for PDCoV M gene, HEX for TGEV N gene, and TexasRed for PEAV N gene. The standard curves for each virus were generated using 1.1 × 107 copies to 1.1 × 101 copies of tenfold serially diluted linearized plasmids conceiving target genes. The results demonstrated that the single real-time RT-qPCR assays for each virus were successfully established at the limit of detection at approximately 10 copies (Fig. 1). All the standard curves showed an excellent correlation coefficient and amplification efficacy, for instance, PEDV (R2 = 1; Eff% = 95.7), PDCoV (R2 = 1; Eff% = 100.9), TGEV (R2 = 0.999; Eff% = 106.0), and PEAV (R2 = 0.995; Eff% = 98.9), indicating that the single real-time RT-qPCR for each virus was valid and reliable.
Fig. 1.
Establishment of single real-time RT-qPCR assay for the individual virus. a The amplification curves (top) and a standard curve (bottom) for detection of PEDV M gene were generated. The probe was labeled with FAM at 5′-end and TAMRA at 3′-end. b The amplification curves (top) and a standard curve (bottom) for detection of PDCoV M gene were generated. The probe was labeled with Cy5 at 5′-end and BHQ2 at 3′-end. c The amplification curves (top) and a standard curve (bottom) for detection of TGEV N gene were generated. The probe was labeled with HEX at 5′-end and BHQ1 at 3′-end. d The amplification curves (top) and a standard curve (bottom) for detection of PEDV N gene were generated. The probe was labeled with TexasRed at 5′-end and BHQ2 at 3′-end
Establishment of multiplex real-time RT-qPCR assay
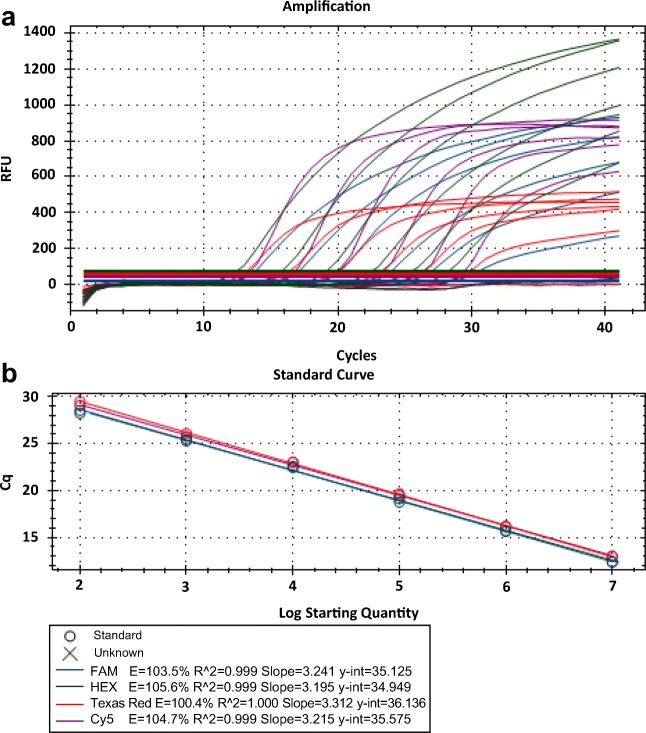
To establish the multiplex real-time RT-qPCR method, all primer sets, probes, and serially diluted standard plasmids for detection of PEDV, PDCoV, TGEV, and PEAV were mixed with 2 × TransStart Probe qPCR SuperMix and nuclease-free water. The concentrations of each primer and probe were optimized for the optimum output. The optimal final concentrations of primers and probes were as follows: 300 nM primer and 100 nM probe for PEDV, 200 nM primer and 200 nM probe for PDCoV, 500 nM primer and 100 nM probe for TGEV, and 500 nM primer and 50 nM probe for PEAV. The results demonstrated that the multiplex real-time RT-qPCR could detect all target genes of these four viruses efficiently with high correlation values (Fig. 2). All the standard curves showed excellent correlation coefficient and amplification efficacy, for details, PEDV (R2 = 0.999; Eff% = 103.5), PDCoV (R2 = 0.999; Eff% = 104.7), TGEV (R2 = 0.999; Eff% = 105.6), and PEAV (R2 = 1; Eff% = 100.4) (Fig. 2). The limit of detection of this multiplex RT-qPCR was approximately 100 copies of each virus per reaction (Fig. 2).
Fig. 2.
Establishment of multiplex real-time RT-qPCR. a Amplification curves and b standard curves of optimized multiplex TaqMan-probe-based real-time RT-qPCR for detection of PEDV, PDCoV, TGEV, and PEAV were generated at the optimum amplification conditions. The correlation coefficient and amplification efficacy of the standards curves, PEDV (R2 = 0.999; Eff% = 103.5), PDCoV (R2 = 0.999; Eff% = 104.7), TGEV (R2 = 0.999; Eff% = 105.6), and PEAV (R2 = 1; Eff% = 100.4), were ideal for detecting the target genes
The specificity of multiplex real-time RT-qPCR assay
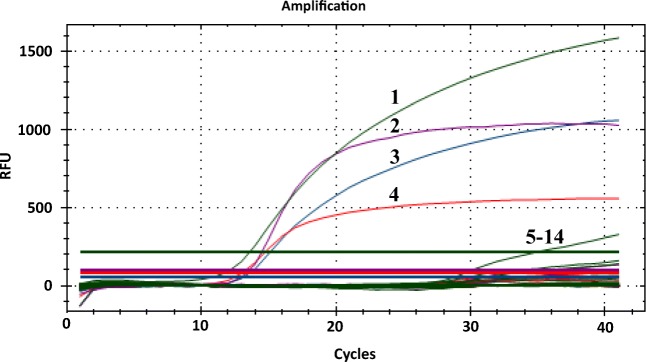
To evaluate the specificity of the multiplex RT-qPCR developed in this study for swine enteric coronavirus detection, the DNAs/cDNAs of nine other major swine viruses were used as templates for amplification with this multiplex system. The cDNAs of PEDV, PDCoV, TGEV, and PEAV were served as positive control while the nuclease-free water was used as a negative control. The results displayed that all swine enteric coronaviruses were successfully detected. However, there was no positive signal detected from other nine swine viruses, PAstV, PKV, FMDV-O, FMDV-A, PRRSV, CSFV, PRV, PCV2, PrV, and negative control using this multiplex RT-qPCR system (Fig. 3), demonstrating that the TaqMan-probe-based multiplex RT-qPCR system was highly specific.
Fig. 3.
The specificity of multiplex TaqMan-probe-based real-time RT-qPCR. 1–4: positive templates of TGEV, PDCoV, PEDV, and PEAV. 5–14: templates of PAsV, PKV, FMDV-O, FMDV-A, CSFV, PRRSV, PRV, PCV2, RrV, and nuclease-free water control
The sensitivity of multiplex real-time RT-qPCR assay
To determine the sensitivity of this multiplex real-time RT-qPCR, tenfold serial dilution of linearized plasmid mixtures were added to the amplification system. The results showed that 1.1 × 102 copies of TGEV, PEDV, PDCoV, and PEAV were detectable with the CT values at 28.75, 28.94, 30.03, and 28.4 respectively (Table 2). However, 1.1 × 101 copies of each target were not detectable in the same amplification system (Table 2). Also, the amplification exhibited reliable and high efficacy, PEDV (R2 = 0.998; Eff% = 96.04), PDCoV (R2 = 0.992; Eff% = 91.38), TGEV (R2 = 0.998; Eff% = 94.57), and PEAV (R2 = 0.999; Eff% = 98.60). This multiplex real-time RT-qPCR demonstrated the same sensitivity level when using viral RNAs as templates (Supplemental Fig. S1). These results implied that the TaqMan-probe-based multiplex real-time RT-qPCR assay could detect porcine enteric coronaviruses (PEDV, TGEV, PDCoV, and PEAV) as less as 10 to 100 copies.
Table 2.
Sensitivity of multiplex RT-qPCR
| Templates (copies/gene) | CT value | |||
|---|---|---|---|---|
| TGEV | PEDV | PDCoV | PEAV | |
| 1.1 × 107 | 11.54 | 12.05 | 12.16 | 11.88 |
| 1.1 × 106 | 15.30 | 15.66 | 16.98 | 15.18 |
| 1.1 × 105 | 18.77 | 18.76 | 20.37 | 18.60 |
| 1.1 × 104 | 22.55 | 22.84 | 24.22 | 22.38 |
| 1.1 × 103 | 25.71 | 26.04 | 27.31 | 25.54 |
| 1.1 × 102 | 28.75 | 28.94 | 30.03 | 28.40 |
| 1.1 × 101 | ND | ND | ND | ND |
| NTC | ND | ND | ND | ND |
Repeatability of multiplex real-time RT-qPCR assay
To estimate the reproducibility of the multiplex real-time PCR, tenfold serial dilution of pooled linearization plasmids were in a triplicate manner for both intra-assay and inter-assay. For intra-assay, the standard plasmids were amplified three times simultaneously. For inter-assay, standard curves were done at three individual times and using a different batch of a standard substance. As shown in Table 3, the coefficient of variation for PEDV intra-assay was 0.30–1.56% and inter-assay was 0.44–1.79%. The PDCoV amplification exhibited the coefficients of variation at 0.09–5.51% and 1.91–3.78% for inter-assay and intra-assay respectively. As for TGEV, the coefficients of variation were 0.33–1.29% and 0.95–3.70% corresponding to intra-assay and inter-assay. The amplification of PEAV showed relative higher coefficients of variation, which was 0.39–7.32% for intra-assay and 1.08–4.52% for inter-assay. These results indicated that the TaqMan-probe-based multiplex real-time RT-qPCR assay established in this study was repeatable and reliable.
Table 3.
Cq value analysis of multiplex real-time RT-qPCR repeatability
| Plasmid | Dilution (copies/μL) | Intra-assay | Inter-assay | ||
|---|---|---|---|---|---|
| Cq value (mean ± SD) | CV% | Cq value (mean ± SD) | CV% | ||
| PEDV | 1.1 × 107 | 12.72 ± 0.15 | 1.18 | 12.31 ± 0.22 | 1.79 |
| 1.1 × 106 | 16.16 ± 0.05 | 0.30 | 15.69 ± 0.07 | 0.44 | |
| 1.1 × 105 | 19.23 ± 0.23 | 1.20 | 18.91 ± 0.25 | 1.32 | |
| 1.1 × 104 | 22.85 ± 0.08 | 0.35 | 22.67 ± 0.15 | 0.66 | |
| 1.1 × 103 | 25.90 ± 0.33 | 1.27 | 25.73 ± 0.40 | 1.55 | |
| 1.1 × 102 | 28.93 ± 0.45 | 1.56 | 28.93 ± 0.37 | 1.29 | |
| 1.1 × 101 | ND | ND | |||
| NTC | ND | ND | |||
| PDCoV | 1.1 × 107 | 12.69 ± 0.70 | 5.51 | 12.71 ± 0.48 | 3.78 |
| 1.1 × 106 | 16.44 ± 0.02 | 0.12 | 16.39 ± 0.52 | 3.17 | |
| 1.1 × 105 | 19.72 ± 0.60 | 3.04 | 19.69 ± 0.60 | 3.05 | |
| 1.1 × 104 | 23.21 ± 0.02 | 0.09 | 23.38 ± 0.73 | 3.12 | |
| 1.1 × 103 | 26.14 ± 0.52 | 2.0 | 26.38 ± 0.82 | 3.11 | |
| 1.1 × 102 | 29.15 ± 0.15 | 0.51 | 29.38 ± 0.56 | 1.91 | |
| 1.1 × 101 | ND | ND | |||
| NTC | ND | ND | |||
| TGEV | 1.1 × 107 | 11.91 ± 0.07 | 0.59 | 11.89 ± 0.44 | 3.70 |
| 1.1 × 106 | 15.10 ± 0.05 | 0.33 | 15.29 ± 0.44 | 2.88 | |
| 1.1 × 105 | 18.32 ± 0.13 | 0.71 | 18.74 ± 0.42 | 2.24 | |
| 1.1 × 104 | 21.86 ± 0.12 | 0.55 | 22.28 ± 0.39 | 1.75 | |
| 1.1 × 103 | 24.98 ± 0.32 | 1.29 | 25.46 ± 0.26 | 1.02 | |
| 1.1 × 102 | 27.83 ± 0.11 | 0.40 | 28.56 ± 0.27 | 0.95 | |
| 1.1 × 101 | ND | ND | |||
| NTC | ND | ND | |||
| PEAV | 1.1 × 107 | 12.16 ± 0.89 | 7.32 | 12.38 ± 0.56 | 4.52 |
| 1.1 × 106 | 15.39 ± 0.1 | 0.65 | 15.57 ± 0.51 | 3.28 | |
| 1.1 × 105 | 18.64 ± 0.05 | 0.27 | 19.11 ± 0.54 | 2.82 | |
| 1.1 × 104 | 22.14 ± 0.07 | 0.32 | 22.60 ± 0.35 | 1.55 | |
| 1.1 × 103 | 25.20 ± 0.29 | 1.15 | 25.85 ± 0.28 | 1.08 | |
| 1.1 × 102 | 28.37 ± 0.11 | 0.39 | 28.97 ± 0.57 | 1.97 | |
| 1.1 × 101 | ND | ND | |||
| NTC | ND | ND | |||
Detection of the clinical samples
The cDNAs originated from 354 clinical samples were subjected to amplification by this multiplex real-time RT-qPCR and confirmed by conventional RT-PCR (Supplemental Fig. S2). The results demonstrated that the samples from Henan and Shaanxi provinces were swine enteric coronavirus–negative while the Gansu province suffered a high prevalence of coronaviruses, showing 83.9% (26/31) TGEV, 48.4% (15/31) PDCoV, and 38.7% (12/31) PEDV positive (Table 4). In Ningxia province, TGEV was also highly prevalent but the positive rates for the other three swine enteric coronaviruses were quite low or not detectable (Table 4). Only PEDV (15/62, 24.2%) and PDCoV (8/62, 12.9%) among these four coronaviruses were detectable from clinical samples of Chongqing. All these four swine enteric coronaviruses were detectable in samples of Liaoning province, with TGEV positive rate at 12.6% (18/143) and the other three were less than 10% (Table 4). Overall, PDCoV (121/354, 34.2%) and TGEV (72/354, 20.3%) were the main coronaviruses detected from all these clinical samples. The positive rate for the newly emerged coronavirus in Guangdong, China, PEAV, was rarely found from these provinces.
Table 4.
Positive rates detected from the clinical samples of different provinces
| Henan | Shaanxi | Liaoning | Gansu | Ningxia | Chongqing | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (n = 63) | (n = 15) | (n = 143) | (n = 31) | (n = 40) | (n = 62) | (n = 354) | ||||||||
| Positive | Percentage (%) | Positive | Percentage (%) | Positive | Percentage (%) | Positive | Percentage (%) | Positive | Percentage (%) | Positive | Percentage (%) | Positive | Percentage (%) | |
| PEDV | 0 | 0 | 0 | 0 | 5 | 3.5% | 12 | 38.7% | 2 | 5% | 15 | 24.2% | 34 | 9.6 |
| PDCoV | 0 | 0 | 0 | 0 | 12 | 8.4% | 15 | 48.4% | 0 | 0% | 8 | 12.9% | 121 | 34.2% |
| TGEV | 0 | 0 | 0 | 0 | 18 | 12.6% | 26 | 83.9% | 28 | 70% | 0 | 0% | 72 | 20.3% |
| PEAV | 0 | 0 | 0 | 0 | 4 | 2.8% | 1 | 3.22% | 0 | 0% | 0 | 0% | 5 | 1.41% |
We further analyzed the co-infection of coronaviruses for all these positive samples. The results elucidated that the main co-infection were caused by a dual coronavirus, for example, 13 PDCoV and TGEV co-infection samples, five PEDV and PDCoV co-infection samples, and four PEDV and TGEV co-infection samples (Fig. 4). Some clinical samples were co-infected by three (9 PEDV, PDCoV, and TGEV co-infection) or all four coronaviruses (Fig. 4). These results indicated the pathogens of viral diarrhea disease in Chinese pig farms were complicated.
Fig. 4.
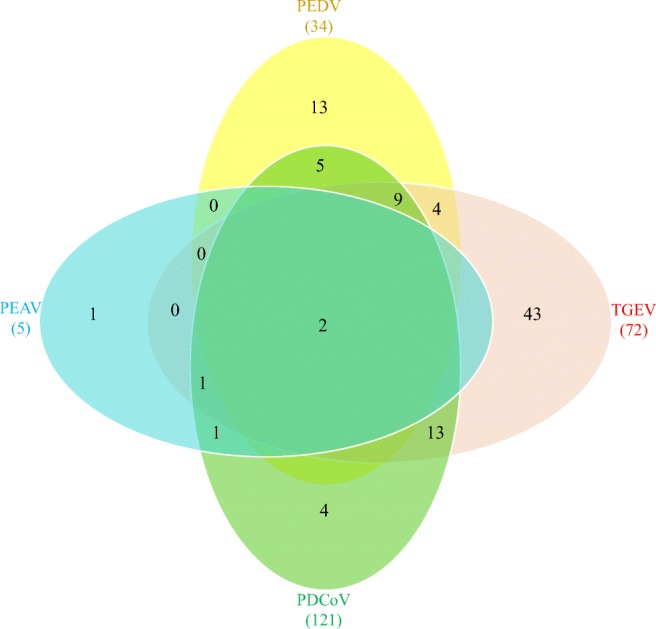
Co-infection analysis between detected swine enteric coronaviruses. A total of 354 clinical diarrhea samples collected from six provinces of China between 2015 and 2018 were subjected to an analysis by the real-time RT-qPCR assay developed in this study. The Venn diagram shows the number of samples infected by either single virus or multi-coronaviruses
Discussion
Porcine diarrheal disease is one of the most severe diseases in pig farms. Swine enteric coronaviruses are the most significant pathogens causing diarrhea in piglets, especially for newborn piglets (Butler et al. 2014). All swine enteric coronaviruses, including traditional PEDV, TGEV, and newly emerged PDCoV, PEAV, could cause serious diarrhea, vomiting, dehydration, weight loss, and up to 100% death in suckling piglets (Gong et al. 2017; Hsu et al. 2018). Previously, some investigators developed a serial of ELISA assays to detect PEDV, TGEV, or PDCoV infections based on M, N proteins or whole virus (Luo et al. 2017; Ma et al. 2016; Su et al. 2016). However, molecular diagnostic tools, rather than serological methods, for swine enteric coronaviruses are urgently needed due to their similar and high pathogenicity to suckling piglets, lacking a mature immune system. For swine enteric coronavirus detection, conventional RT-PCR, multiplex RT-qPCR, or pan-coronaviruses RT-PCR methods targeting M, N, S, or polymerase genes were developed in recent years (Hsu et al. 2018; Song et al. 2006). Additionally, Zhou et al. (2018a) recently reported a specific real-time PCR for the detection of PEAV. To our knowledge, there was no such method that could simultaneously detect and differentiate TGEV, PEDV, PDCoV, and PEAV from the same detection vial. To solve this urgent and important issue, we developed this multiplex real-time RT-qPCR for the detection and differential diagnosis of the swine enteric coronaviruses circulating in pig herds.
TaqMan-probe-based real-time qPCR is a rapid, high specificity, high sensitivity, and great reproducibility detection tool for identification of viruses (Chang et al. 2014). Among the seven open reading frames (ORFs) and four structural protein genes, S (spike), E (envelope), M (membrane), N (nucleocapsid) of coronaviruses (Su et al. 2018), the M and N genes are highly conserved within the same antigenic group but less homologous between each of these four swine enteric coronaviruses (Yang et al. 2019). To establish a high-specific multiplex real-time RT-qPCR for the detection and differential diagnosis of swine enteric coronaviruses, primer sets, and TaqMan probes were designed targeting the highly conserved regions of PEDV or PDCoV M genes and TGEV or PEAV N genes, based on bioinformatics analysis of each virus. Our results demonstrated that each primer and probe set can only detect target gene itself and could not bind with any other targets, indicating high specificity.
Our results showed that the singular real-time RT-qPCRs were capable to detect as less as 10 copies of PEDV, PDCoV, TGEV, and PEAV templates. However, the detection limit of each target gene in the multiplex real-time RT-qPCR was approximately 100 copies, implying that the sensitivity of multiplex real-time RT-qPCR was lower than that of singular real-time RT-qPCR probably due to the competition between primers, probes, templates, and reagents.
The analysis of clinical diarrhea samples using this developed multiplex RT-qPCR elucidated that the co-infection of swine enteric coronaviruses commonly existed in some pig farms. Co-infection of swine enteric coronaviruses may cause recombination between co-infected viruses. During 2012 and 2016, some new swine enteric coronaviruses were generated by recombination with PEDV and TGEV and spread across the central Eastern European countries (Akimkin et al. 2016; Belsham et al. 2016; Boniotti et al. 2016). The recombination might create more virulent enteric virus strains or new viruses, leading to potential outbreaks or pandemics of swine viral diarrhea. Additionally, co-infections may promote the evolution of non-pathogenic enteric viruses into high virulent and pathogenic viruses. A recent example showed the existence of PDCoV in pig herds for many years in China mainland and Hong Kong but without observed clinical symptoms (Pan et al. 2017; Woo et al. 2012). Unfortunately, this virus caused severe outbreaks in some states of the USA since 2014 (Ma et al. 2015; Wang et al. 2014). This virulence change was most probably caused by the previous exposure to PEDV or other co-infected swine enteric viruses. Therefore, swine enteric coronaviruses co-infection may bring risks for the prevention and control of swine diarrhea.
In summary, we developed a TaqMan-probe-based multiplex real-time RT-qPCR with high specificity and sensitivity for simultaneous detection and differential diagnosis of swine enteric coronaviruses, PEDV, TGEV, PDCoV, and PEAV. This real-time RT-qPCR assay is of great significance for the prevention and control and epidemiological investigation of swine viral diarrhea.
Electronic supplementary material
(PDF 149 kb)
Funding information
This study was supported by the National Key R&D Program of China (2016YFD0500103), National Natural Science Foundation of China (31572498, 31702209), the Elite Youth Program of CAAS, and partly by China Central Public-interest Scientific Institution Basal Research Fund (1610312018002).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jinquan Wang, Email: Wangjinquan163@163.com.
Guangliang Liu, Phone: +86(931)834-2682, Email: LiuGuangliang01@caas.cn.
References
- Akimkin V, Beer M, Blome S, Hanke D, Hoper D, Jenckel M, Pohlmann A. New chimeric porcine coronavirus in swine feces, Germany, 2012. Emerg Infect Dis. 2016;22(7):1314–1315. doi: 10.3201/eid2207.160179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham GJ, Rasmussen TB, Normann P, Vaclavek P, Strandbygaard B, Botner A. Characterization of a novel chimeric swine enteric coronavirus from diseased pigs in central eastern Europe in 2016. Transbound Emerg Dis. 2016;63(6):595–601. doi: 10.1111/tbed.12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniotti MB, Papetti A, Lavazza A, Alborali G, Sozzi E, Chiapponi C, Faccini S, Bonilauri P, Cordioli P, Marthaler D. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg Infect Dis. 2016;22(1):83–87. doi: 10.3201/eid2201.150544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE, Lager KM, Golde W, Faaberg KS, Sinkora M, Loving C, Zhang YI. Porcine reproductive and respiratory syndrome (PRRS): an immune dysregulatory pandemic. Immunol Res. 2014;59(1–3):81–108. doi: 10.1007/s12026-014-8549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, Deng MC, Wang FI, Tsai HJ, Yang CH, Chang C, Huang YL. The application of a duplex reverse transcription real-time PCR for the surveillance of porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. J Virol Methods. 2014;201:13–19. doi: 10.1016/j.jviromet.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Gong L, Li J, Zhou Q, Xu Z, Chen L, Zhang Y, Xue C, Wen Z, Cao Y (2017) A new bat-HKU2-like coronavirus in swine, China, 2017. Emerg Infect Dis 23(9):1607–1609. 10.3201/eid2309.170915 [DOI] [PMC free article] [PubMed]
- Hsu TH, Liu HP, Chin CY, Wang C, Zhu WZ, Wu BL, Chang YC. Detection, sequence analysis, and antibody prevalence of porcine deltacoronavirus in Taiwan. Arch Virol. 2018;163:3113–3117. doi: 10.1007/s00705-018-3964-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YL, Pang VF, Pan CH, Chen TH, Jong MH, Huang TS, Jeng CR. Development of a reverse transcription multiplex real-time PCR for the detection and genotyping of classical swine fever virus. J Virol Methods. 2009;160(1–2):111–118. doi: 10.1016/j.jviromet.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo SX, Fan JH, Opriessnig T, Di JM, Liu BJ, Zuo YZ. Development and application of a recombinant M protein-based indirect ELISA for the detection of porcine deltacoronavirus IgG antibodies. J Virol Methods. 2017;249:76–78. doi: 10.1016/j.jviromet.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyoo KS, Yeom M, Kim J, Kim D, Ha G, Na W, Le VP, Song D. Development of rapid immunochromatographic strip test for the detection of porcine epidemic diarrhoea virus. Vet Rec. 2017;181(22):596–601. doi: 10.1136/vr.103959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhang Y, Liang X, Lou F, Oglesbee M, Krakowka S, Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio. 2015;6(2):e00064. doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhang Y, Liang X, Oglesbee M, Krakowka S, Niehaus A, Wang G, Jia A, Song H, Li J. Two-way antigenic cross-reactivity between porcine epidemic diarrhea virus and porcine deltacoronavirus. Vet Microbiol. 2016;186:90–96. doi: 10.1016/j.vetmic.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Tian X, Qin P, Wang B, Zhao P, Yang YL, Wang L, Wang D, Song Y, Zhang X, Huang YW. Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet Microbiol. 2017;211:15–21. doi: 10.1016/j.vetmic.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song DS, Kang BK, Oh JS, Ha GW, Yang JS, Moon HJ, Jang YS, Park BK. Multiplex reverse transcription-PCR for rapid differential detection of porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine group a rotavirus. J Vet Diagn Investig. 2006;18(3):278–281. doi: 10.1177/104063870601800309. [DOI] [PubMed] [Google Scholar]
- Song D, Zhou X, Peng Q, Chen Y, Zhang F, Huang T, Zhang T, Li A, Huang D, Wu Q, He H, Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound Emerg Dis. 2015;62(6):575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M, Li C, Guo D, Wei S, Wang X, Geng Y, Yao S, Gao J, Wang E, Zhao X, Wang Z, Wang J, Wu R, Feng L, Sun D. A recombinant nucleocapsid protein-based indirect enzyme-linked immunosorbent assay to detect antibodies against porcine deltacoronavirus. J Vet Med Sci. 2016;78(4):601–606. doi: 10.1292/jvms.15-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Liu Y, Chen Y, Xing G, Hao H, Wei Q, Liang Y, Xie W, Li D, Huang H, Deng R, Zhang G. A novel duplex TaqMan probe-based real-time RT-qPCR for detecting and differentiating classical and variant porcine epidemic diarrhea viruses. Mol Cell Probes. 2018;37:6–11. doi: 10.1016/j.mcp.2017.10.003. [DOI] [PubMed] [Google Scholar]
- Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, Song CX. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis. 2012;18(1):161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Byrum B, Zhang Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis. 2014;20(7):1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Huang Y, Lau SK, Yuen KY. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chen W, Huang J, Jin L, Zhou Y, Chen J, Zhang N, Wu D, Sun E, Liu G (2019) Generation, identification and functional analysis of monoclonal antibodies against porcine epidemic diarrhea virus nucleocapsid. Appl Microbiol Biotechnol 103: 3705-3714. 10.1007/s00253-019-09702-5 [DOI] [PMC free article] [PubMed]
- Zhang J, Tsai YL, Lee PY, Chen Q, Zhang Y, Chiang CJ, Shen YH, Li FC, Chang HF, Gauger PC, Harmon KM, Wang HT. Evaluation of two singleplex reverse transcription-insulated isothermal PCR tests and a duplex real-time RT-PCR test for the detection of porcine epidemic diarrhea virus and porcine deltacoronavirus. J Virol Methods. 2016;234:34–42. doi: 10.1016/j.jviromet.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Sun Y, Wu JL, Mai KJ, Chen GH, Wu ZX, Bai Y, Li D, Zhou ZH, Cheng J, Wu RT, Zhang XB, Ma JY. Development of a TaqMan-based real-time RT-PCR assay for the detection of SADS-CoV associated with severe diarrhea disease in pigs. J Virol Methods. 2018;255:66–70. doi: 10.1016/j.jviromet.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, Zhu Y, Zhang YW, Xie QM, Mani S, Zheng XS, Li B, Li JM, Guo H, Pei GQ, An XP, Chen JW, Zhou L, Mai KJ, Wu ZX, Li D, Anderson DE, Zhang LB, Li SY, Mi ZQ, He TT, Cong F, Guo PJ, Huang R, Luo Y, Liu XL, Chen J, Huang Y, Sun Q, Zhang XL, Wang YY, Xing SZ, Chen YS, Sun Y, Li J, Daszak P, Wang LF, Shi ZL, Tong YG, Ma JY. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556(7700):255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 149 kb)