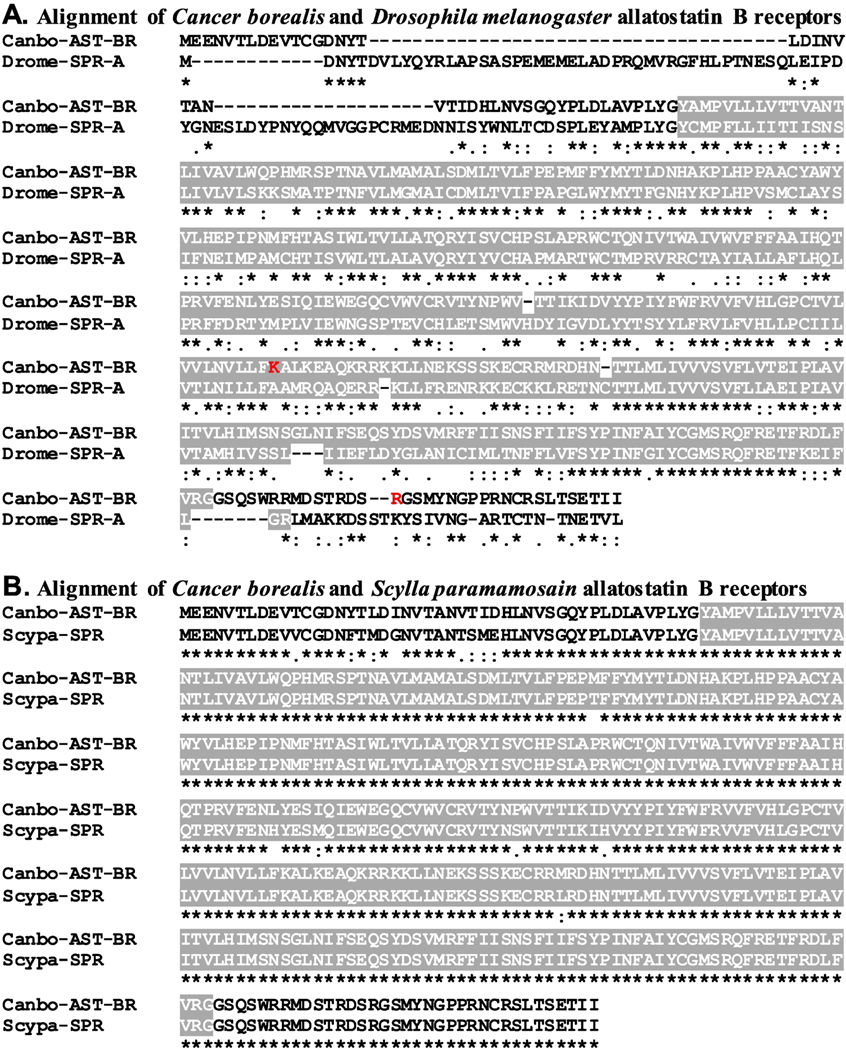
Figure 3.
Alignment of Cancer borealis and related allatostatin B/sex peptide receptors. (A) MAFFT alignment of the putative C. borealis allatostatin B receptor (Canbo-AST-BR; deduced from GEFB01014490) and the Drosophila melanogaster sex peptide receptor (Drome-SPR; Accession No. AAF46037). (B) MAFFT alignment of Canbo-AST-BR and the Scylla paramamosain sex peptide receptor (Scypa-SPR; Accession No. ANF04993). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “.” denote amino acids that are similar in structure between sequences. In this figure, serpentine receptor class W seven-transmembrane domains identified by Pfam analyses are highlighted in gray. In A, the residues that vary between the transcriptome derived Canbo-AST-BR sequence and that deduced from the cloned transcript MH729783, i.e., Lys250 to Arg and Arg378 to His, are shown in red font.