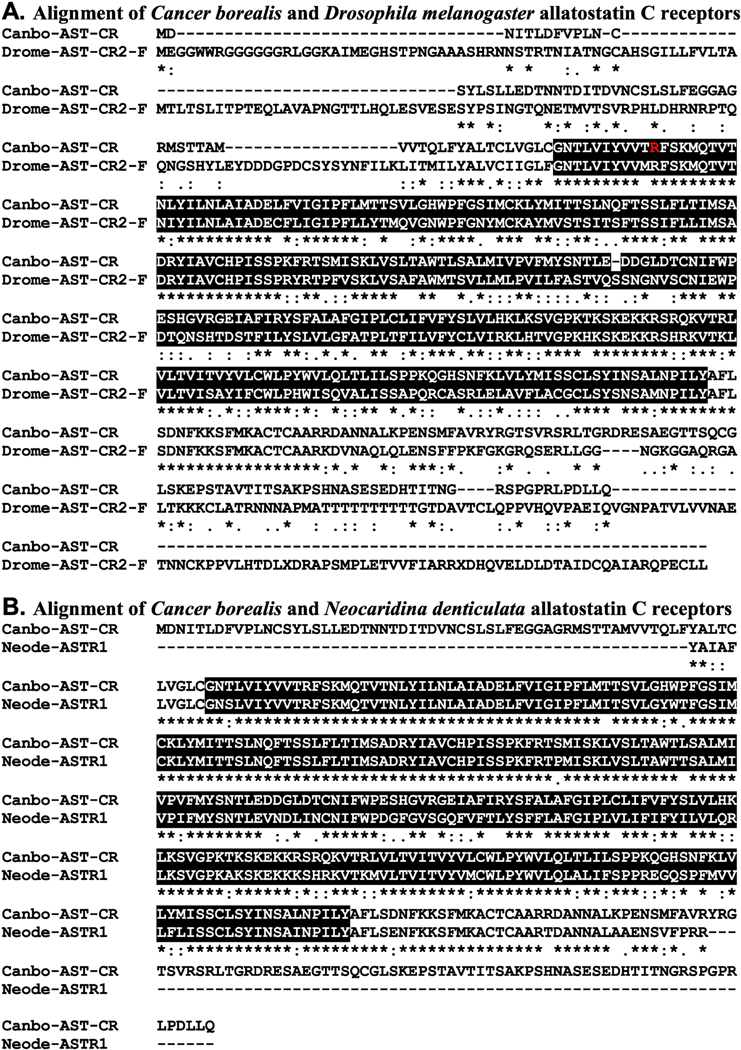
Figure 4.
Alignment of Cancer borealis and related allatostatin C receptors. (A) MAFFT alignment of the putative C. borealis allatostatin C receptor (Canbo-AST-CR; deduced from GEFB01019215) and Drosophila melanogaster allatostatin C receptor 2, isoform F (Drome-AST-CR2-F; Accession No. ALI30485). (B) MAFFT alignment of Canbo-AST-CR and the extant portion of Neocaridina denticulata allatostatin receptor 1 (Neode-ASTR1; Accession No. AIY69136). In the line immediately below each sequence grouping, “*” indicates identical amino acid residues, while “:” and “” denote amino acids that are similar in structure between sequences. In this figure, rhodopsin family seven-transmembrane receptor domains identified by Pfam analyses are highlighted in black. In A, the residue that varies between the transcriptome derived Canbo-AST-CR sequence and that deduced from the cloned transcript MH729784, i.e., Arg76 to Gly, is shown in red font.