Figure 6.
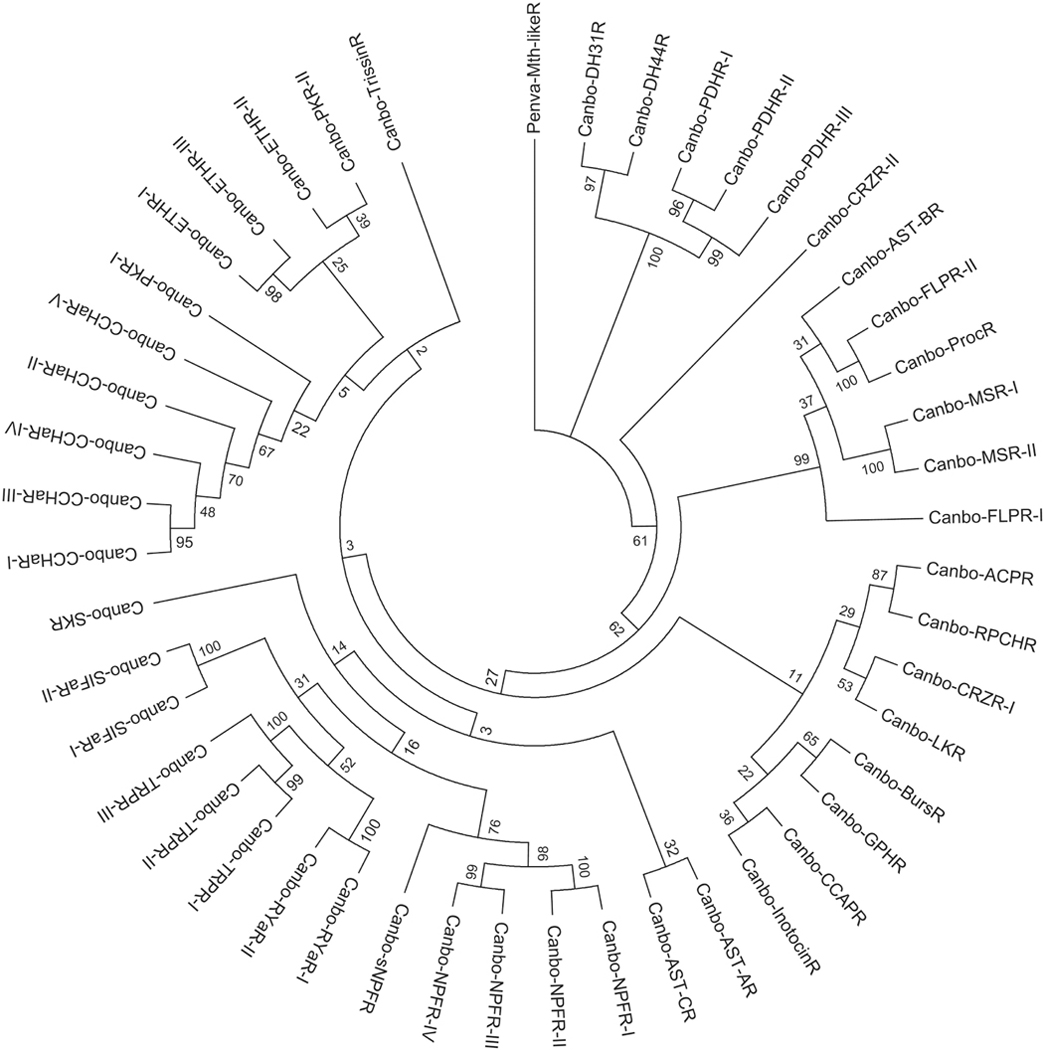
Phylogenetic relationships among putative Cancer borealis peptide receptors. Neighbor-joining tree depicting relationships among the 46 C. borealis peptide receptor sequences identified from mixed neural tissue transcriptomic data. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Abbreviations are as follows: ACPR (adipokinetic hormone-corazonin-like peptide receptor); AST-AR (allatostatin A receptor); AST-BR (allatostatin B receptor); AST-CR (allatostatin C receptor); BursR (bursicon receptor); CCHaR (CCHamide receptor); CRZR (corazonin receptor); CCAPR (crustacean cardioactive peptide receptor); DH31R (diuretic hormone 31 receptor); DH44R (diuretic hormone 44 receptor); ETHR (ecdysis-triggering hormone receptor); FLPR (FMRFamide-like peptide receptor); GPHR (glycoprotein hormone receptor); InotocinR (inotocin receptor); LKR (leucokinin receptor); MSR (myosuppressin receptor); NPFR (neuropeptide F receptor); PDHR (pigment dispersing hormone receptor); ProcR (proctolin receptor); PKR (pyrokinin receptor); RPCHR (red pigment concentrating hormone receptor); RYaR (RYamide receptor); sNPFR (short neuropeptide F receptor); SIFaR (SIFamide receptor); SKR (sulfakinin receptor); TRPR (tachykinin-related peptide receptor); TrissinR (trissin receptor). The tree was rooted using the Penaeus vannamei Methuselah-like 1 GPCR sequence (Penva-Mth-likeR; Accession No. XP 027232612) as an outgroup.
