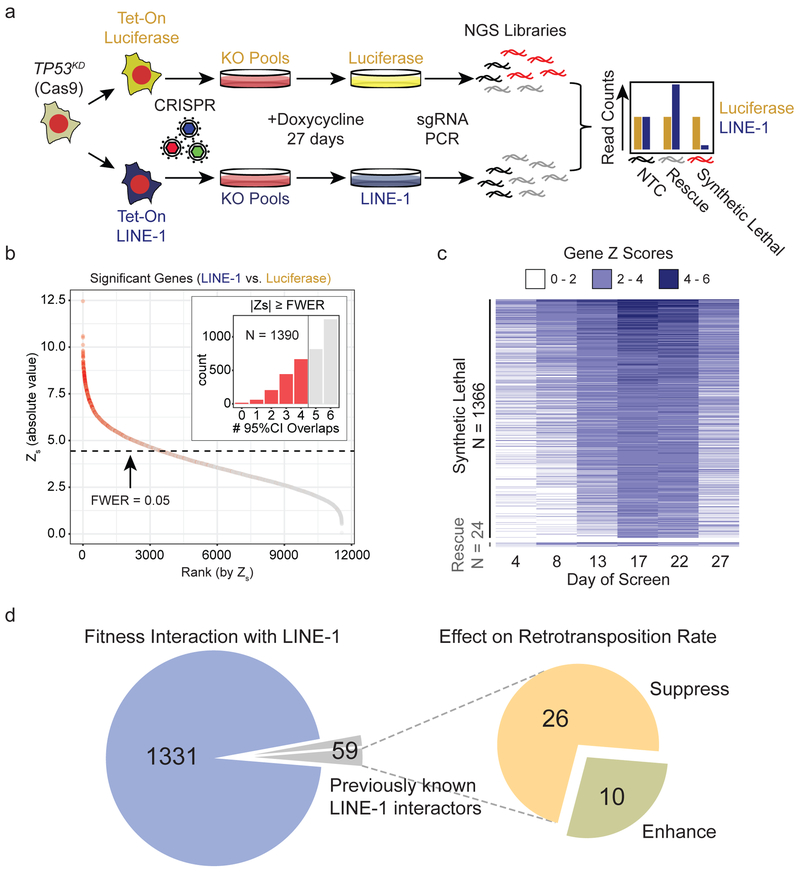
Figure 4. Mapping LINE-1 fitness interactions in TP53-deficient cells.
(a) TP53KD cells are RPE-Cas9 cells stably transduced with shRNA to knockdown p53 and then engineered to express luciferase (pDA094) or codon-optimized LINE-1 (pDA095) in a doxycycline-inducible manner (Tet-On). Tet-On cells were transduced with the Brunello CRISPR KO library at a multiplicity of infection of 0.3 and puromycin-selected for 8 days before inducing expression of LINE-1 or luciferase for 27 days. Cell pools were sampled at 4-5 day intervals and analyzed for sgRNA representation with MAGeCK. Count data are normalized to reads that align to 1,000 built-in non-targeting-control (NTC) sgRNAs (black). NGS = Next Generation Sequencing. KO = Knockout. (b) Genes shown as rank ordered plot of Stauffer Z scores (Zs) with a family-wise error rate (FWER) of 0.05. Inset indicates the number of 95% confidence interval overlaps over all time points between LINE-1 and luciferase groups among gene knockouts that meet the FWER threshold (red) versus those that do not (gray). (c) Heatmap of 1,390 significant genes depicting the Z scores over time, ranked by Zs. There are 1,366 synthetic lethal interactions and 24 rescue interactions. Most knockouts achieved detectable effects by 17-22 days into the screen, evidenced by increasing gene Z scores during these time points. (d) Overlap of genes with LINE-1 fitness interactions observed in the present study with genes previously known to interact with LINE-1 proteins physically or by modifying retrotransposition. Previously known LINE-1 interactors were identified by Liu et al., 2018, Moldovan et al., 2015, Taylor et al., 2013, and Goodier et al., 2013.