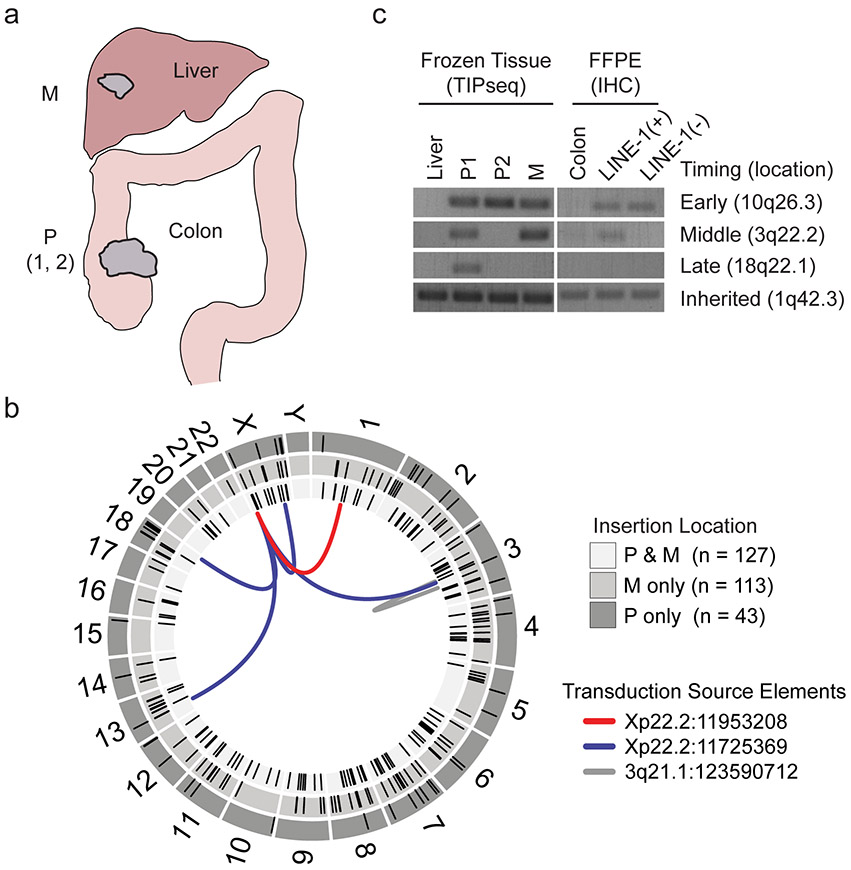
Extended Data Fig. 1. LINE-1 heterogeneity in colon cancer.
(a) Tissues collected for transposon insertion profiling by sequencing (TIP-seq) mapping of tumor-specific LINE insertions. Fresh frozen tissue was collected from two sites in the primary tumor in the colon and one site in the metastatic tumor in the liver. Normal tissue was collected from the liver. The liver metastasis exhibited ORF1p immunoreactivity as well (data not shown). (b) Circos plot detailing TIP-seq results and whether insertions were found in the primary (P only), metastasis (M only) or in both (P & M). In the validation process, we identified 11 3’ transduction events, 6 of which mapped to two LINE-1 sequences on Xp22.2 and one on 3q21.1 that are known to be highly active tumor alleles. As expected, the majority of this tumor’s de novo insertions were intronic or intergenic and not near known tumor suppressors or oncogenes. (c) We genotyped the insertions using hemi-specific PCR in genomic DNA obtained from dissected histology slides and compared to the allele’s presence in bulk frozen tissue used for TIP-seq. In all samples, we detected an inherited LINE-1 on 1q42.3, indicating that our PCR conditions were sufficient to detect LINE-1 elements. An early de novo insertion on 10q26.3 was found in all frozen tissue samples (primary and metastasis) and both CDX2-high and CDX2-dim slide-dissected samples. An insertion on 3q22.2 is present in the primary tumor subclonally and in the metastasis and therefore occurred before metastasis but after dedifferentiation of the CDX2-dim clone. An insertion on 18q22.1 occurred after metastasis to the liver had occurred, since it was found in the primary CDX2-high clone and not in the metastasis.