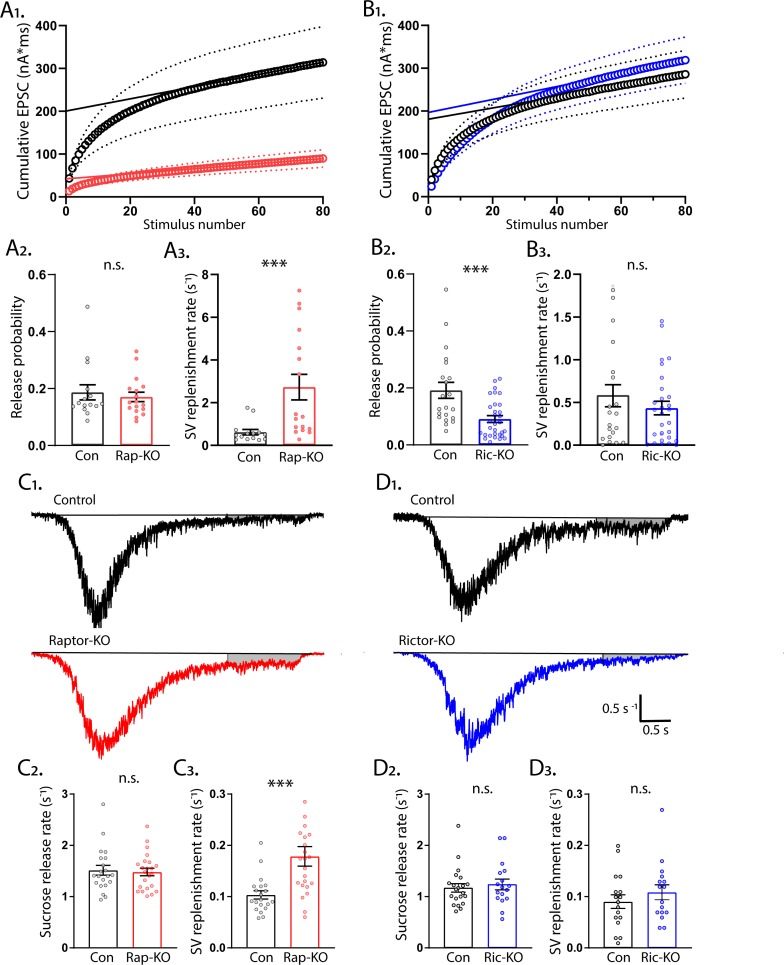
Figure 7. mTORC1 inactivation increases SV replenishment rate.
(A1) Cumulative EPSC charge in Raptor-Con (black) and Raptor-KO (red) neurons. Circles are the population means and the dotted lines are the s.e.m. The solid lines are linear fits to the final 20 responses and the y-intercept indicates the RRPtrain. (A2) Release probability calculated using RRPtrain is unchanged in Raptor-KO neurons. (A3) The SV replenishment rate is significantly increased by Raptor loss. (B1) Cumulative EPSC charge in Rictor-Con (black) and Rictor-KO (blue) neurons. (B2) Release probability calculated using RRPtrain is significantly decreased in Rictor-KO neurons. (B3) The SV replenishment rate is unaffected by Rictor loss. (C1) Example traces of normalized current responses to 500 mM sucrose application in Raptor-Con (black) and Raptor-KO (red) neurons. The black line shows the pre-sucrose baseline, and the gray shaded area shows the area used to calculate the replenishment rate. (C2) Plot showing the rate constants for sucrose-induced synaptic vesicle (SV) release in Raptor-Con (black) and Raptor-KO (red) neurons. (C3) Plot showing the replenishment rate constants after sucrose-induced SV release in Raptor-Con (black) and Raptor-KO (red) neurons. (D1) Example traces of normalized current responses to 500 mM sucrose application in Rictor-Con (black) and Rictor-KO (blue) neurons. The black line shows the pre-sucrose baseline, and the gray shaded area shows the area used to calculate the replenishment rate. (D2) Plot showing the rate constants for sucrose-induced SV release in Rictor-Con (black) and Rictor-KO (blue) neurons. (D3) Plot showing the replenishment rate constants after sucrose-induced SV release in Rictor-Con (black) and Rictor-KO (blue) neurons. In the dot plots, each dot represents the mean value from one neuron obtained from three independent cultures per genotype, and the bars show the estimated marginal means and s.e.m. ***=p < 0.001, and n.s. = p > 0.05, as tested with Generalized Estimating Equations.