Fig. 1. Schematic illustration of our approach to distinguish cell types.
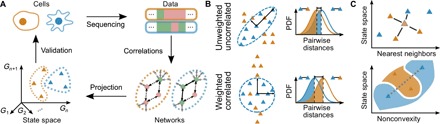
(A) The epigenomic measurements of two different cells in blue and orange (top left) yield different epigenotypes (top right) from which an condition-specific effective network (bottom right) is determined from correlations in the data, where solid or dashed lines indicate relationships that are enforced or not enforced but possible, respectively, under the specified conditions. Projection to the state space of correlation eigenvectors approximates the attractors. (B) The probability distribution functions of distances between pairs of measurements of the same and different types are compared at selected percentiles (shaded regions) to determine whether pairs of the same type are more similar than pairs of different types. (C) The performance is evaluated by using KNN to predict unseen data (top) and by measuring the frequency with which chords cross cell type boundaries (gray dashed line, bottom panel).
