Figure 1.
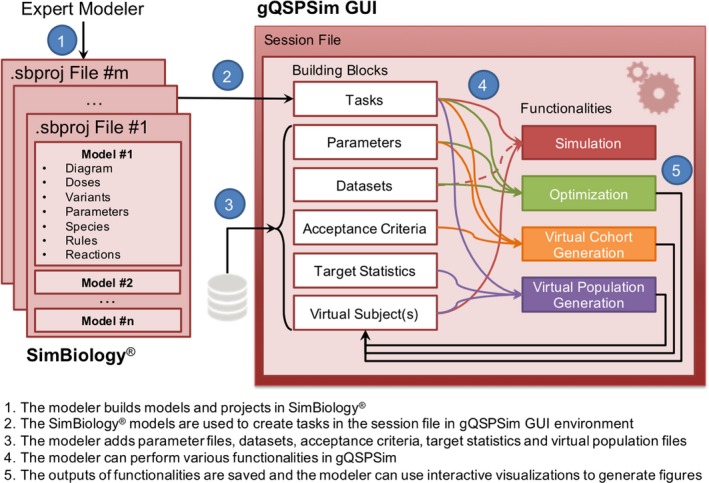
A schematic workflow of how modelers employ SimBiology and gQSPSim. An expert modeler builds the model diagram, variants, dosing objects, and so on in SimBiology. Simultaneously, the modeler can launch a new session in gQSPSim and use SimBiology models to create tasks with the settings specified in the Methods section. The modeler can also add parameter files, datasets, acceptance criteria, target statistics, and virtual subjects as building blocks to perform a variety of functionalities including simulation, Optimization, Virtual Cohort Generation, and Virtual Population Generation. Because the session file only stores the folder path to model files and datasets and does not store their content, the modeler can change models and files outside of gQSPSim, save the files, and seamlessly run the previously defined functionalities. All of the functionalities are supported by features such as interactive visualization and export of results as presentation‐ready figures and Excel files. The session file can be saved for future use. There is an option to autosave the session files and also to use parallel processing to speed up the execution of functionalities. GUI, graphical user interface.
