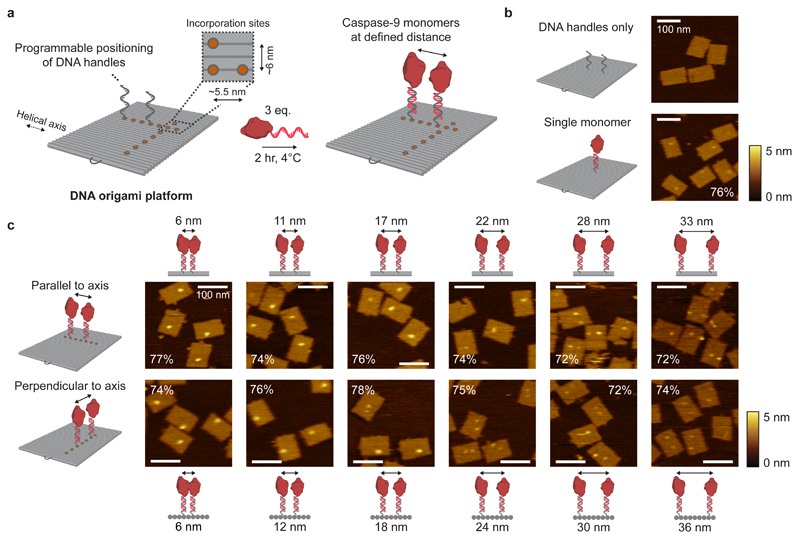
Figure 2. Characterisation of caspase-9 assembly onto DNA origami nanostructures.
a, Schematic of the general strategy for caspase-9 incorporation on DNA origami platforms. By including appropriate handle-extended staple strands during the self-assembly process the position of, and distance between, two ssDNA handles can be controlled. Specifically, the DNA origami technique enables a minimal distance between incorporation sites (orange circles) of 5.5 nm parallel, and 6 nm perpendicular to the DNA helical axis (Supplementary Methods (Self-assembly of DNA origami nanostructures)). Incubation of complementary enzyme-DNA conjugates leads to hybridization and incorporation of two caspase-9 monomers at defined distances. Typically, 4 nM DNA origami was incubated with 3 equivalents of enzyme-DNA conjugate per handle for 2 hr at 4°C. For AFM imaging, functionalized nanostructures were purified using 1.5% agarose gel extraction. b,c, Topographic AFM (tapping mode in solution) images of control samples (b) and DNA origami nanostructures functionalized with two 32-kDa caspase-9 monomers (c) at various distances parallel (top row) and perpendicular (bottom row) to the helical axis. The caspase-9 incorporation efficiency per handle is indicated in percentages, and was calculated based on at least 250 well-formed nanostructures using 4 different images per sample. Colour bars indicate height scale in AFM images. Scale bars, 100 nm.