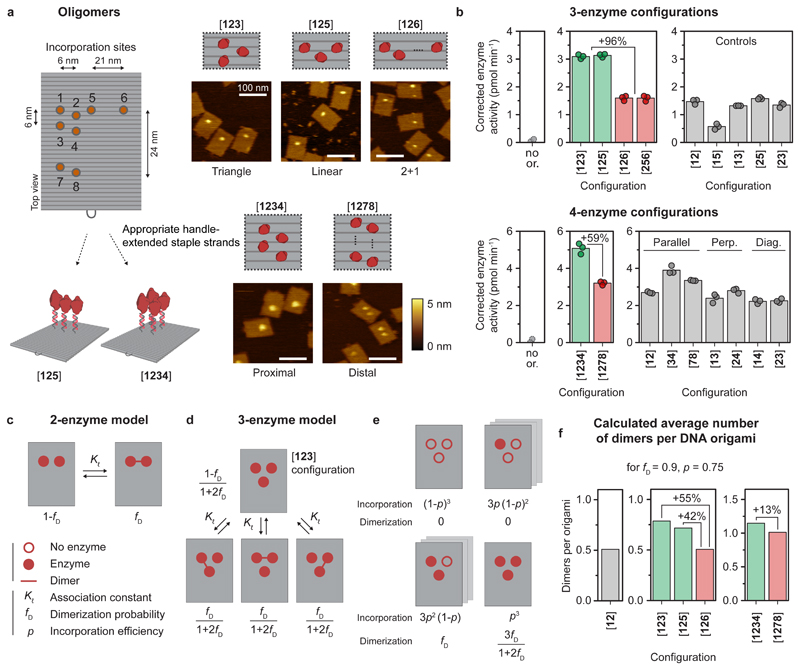
Figure 4. Co-localization of more than two caspase-9 monomers leads to enhanced enzymatic activity.
Reactions were performed as described in Fig. 3. The DNA origami concentration was adjusted to keep the total concentration of enzyme at 24 nM (see Methods and Supplementary Table 3). All experiments were performed in triplicate. a, Schematic overview of possible incorporation sites for handle-extended staple strands (orange circles) for constructing three- and four-enzyme DNA nanostructures. Topographic AFM images show successful incorporation of caspase-9 according to the pre-programmed positions. The bracket notation indicates enzyme configuration as determined by the number and location of the indicated incorporation sites. Colour bars indicate height scale. Scale bars, 100 nm. b Enzymatic activity measurements for three-enzyme (top) and four-enzyme (bottom) DNA nanostructures (green and red) and two-enzyme controls (gray). Activity was corrected by subtracting the mean background activity (no or.) in all samples. Labels: no or., no DNA origami present; perp., perpendicular arrangement; diag., diagonal arrangement. c, Schematic depiction of the mass-balance model for a tethered two-enzyme system, with an equilibrium between a monomeric (left) and dimeric (right) state defined by effective association constant Kℓ, leading to the dimerization probability fD. d, Model for the tethered three-enzyme system in triangular configuration ([123]) (see Supplementary Note 1 for models of other configurations). One monomeric and three symmetric dimeric states can be defined, with corresponding state fractions expressed as a function of fD. e, Schematic depiction of the possible configurations in the triangular three-enzyme system with incomplete enzyme incorporation. f, When taking into account all possible states and incomplete incorporation, the models predict the average number of enzyme dimers per DNA origami for all two-, three-, and four-enzyme systems.