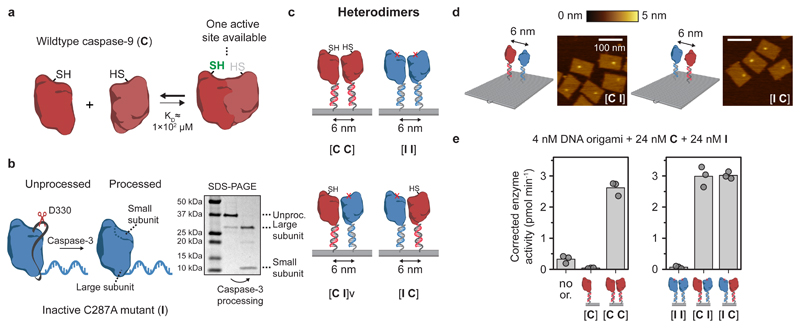
Figure 5. Enzymatic activity of the caspase-9 dimer originates from a single catalytic site.
a, Schematic overview of wildtype caspase-9 (C) homodimerization. Crystal structures of dimeric caspase-9 indicate that only one of the two active sites is in an active, accessible conformation. b, Wildtype caspase-9 undergoes autoproteolytic processing at three cleavage sites in the intersubunit linker region (black ribbon), but mutant C287A (I) is inactive and therefore does not undergo autoprocessing. To mimic processing, enzyme-DNA conjugates were incubated with caspase-3, a constitutively active protease that also cleaves caspase-9 in the linker region. Purification was performed as described in Fig. 1, and confirmed by SDS-PAGE under reducing conditions. Label: unproc., unprocessed enzyme-DNA conjugate. c, The modularity of DNA origami was exploited to construct both homo- and heterodimeric 6-nm two-enzyme DNA nanostructures consisting of combinations of wildtype and inactive monomers. The specific configuration of wildtype (C) and inactive (I) mutants is denoted using bracket notation. d, Topographic AFM images of [I C] and [C I] heterodimers. Colour bar indicates height scale. Scale bars, 100 nm. e, Enzymatic activity measurements were performed as described in Fig. 3. In all samples, both 24 nM C and 24 nM processed I were added (for data with unprocessed I, see Supplementary Fig. 26). Activity was corrected by subtracting the mean background activity in all samples. All experiments were performed in triplicate. Label: no or., no DNA origami present.