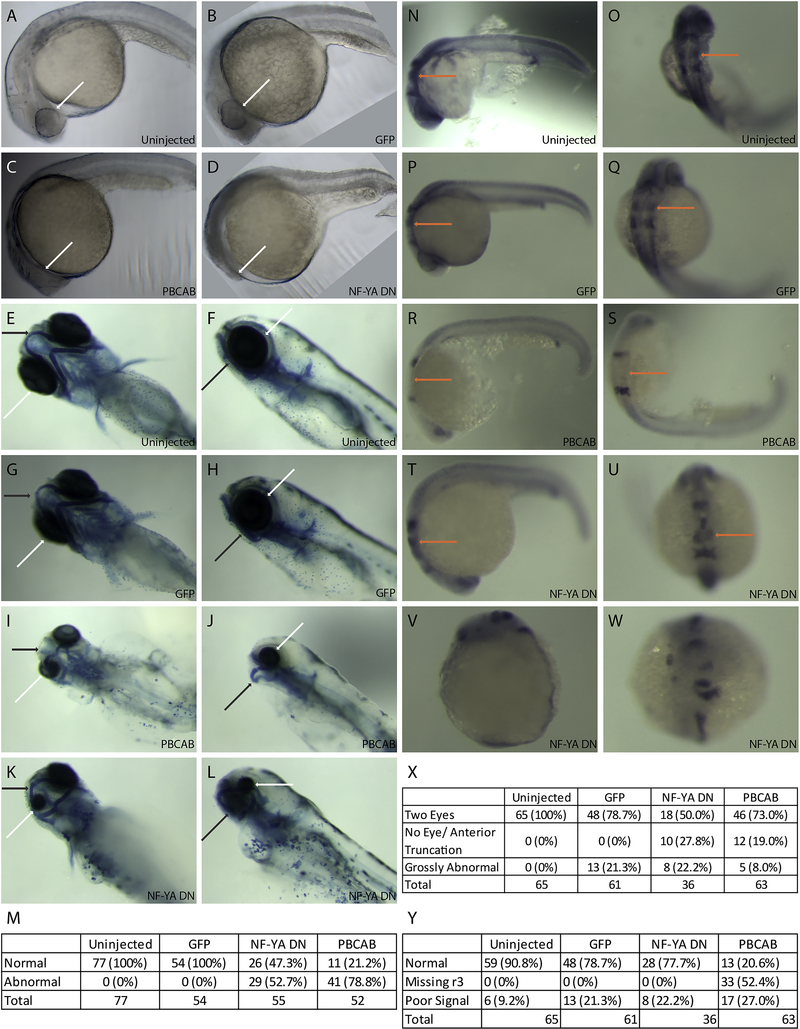
Figure 1: Disruption of NF-Y or TALE function affects anterior embryonic development.
Zebrafish embryos were left uninjected (A, E, F, N, O) or injected with either control mRNA (GFP; B, G, H, P, Q), mRNA encoding a TALE dominant negative construct (PBCAB; C, I, J, R, S) or mRNA encoding an NF-Y dominant negative construct (NF-YA DN; D, K, L, T-W) at the 1–2 cell stage and raised to 24hpf (N-W), 28hpf (A-D) or 5dpf (E-L). Embryos were either left untreated (A-D), stained with alcian blue (E-L) or processed for detection of pax2 (at the mid/hindbrain boundary), krox20 (in rhombomeres 3 and 5) and hoxd4 (in the spinal cord) transcripts by in situ hybridization (N-W). White arrows highlight differences in eye morphology (A-L), black arrows highlight differences in head cartilage formation (E-L) and orange arrows indicate differences in rhombomere 3 krox20 expression (N-U). Tables summarize effects of TALE or NF-Y disruption on head cartilage formation (M), eye formation (X) and gene expression (Y). Panels V and W show representative images of embryos scored as having gross abnormalities in panel X. Embryos are shown in lateral (A-D, F, H, J, L, N, P, R, T, V) or dorsal (E, G, I, K, O, Q, S, U, W) views.