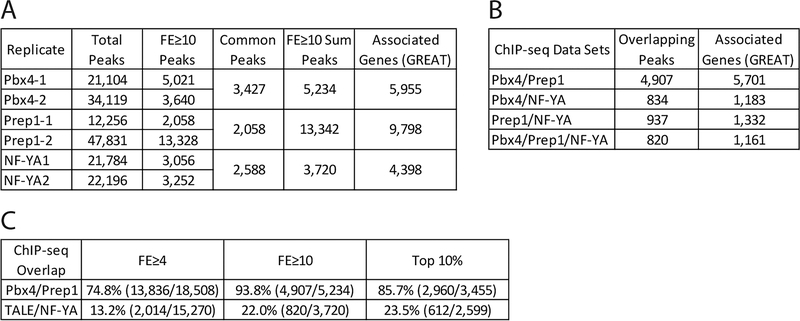
Figure 5: Identification of genomic binding sites for NF-Y and Pbx4 in 3.5hpf zebrafish.
(A) Table showing data for Pbx4 and NF-YA ChIP-seq biological replicates with our previous Prep1 ChIP-seq data REF included as reference. (B) Table showing number of peaks that overlap (at least 1bp shared between 200bp fragments centered on peaks) between Prep1, Pbx4 and NF-YA ChIP-seq data sets. Only peaks with a 10-fold or greater enrichment over input were considered. (C) Table showing extent of overlap of Pbx4 peaks with Prep1 peaks and TALE peaks with NF-YA peaks at three different cutoffs (FE≥4, FE≥10 and top 10% of peaks).