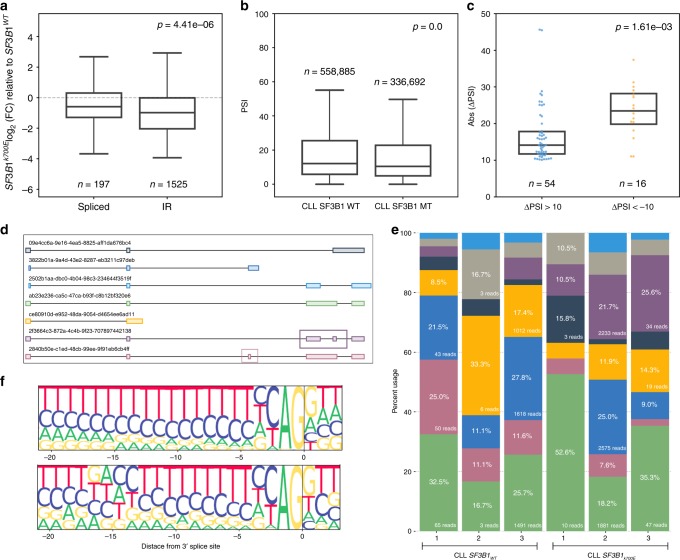
Fig. 4. Intron retention events are more strongly downregulated in CLL SF3B1K700E.
a Expression fold-change between SF3B1K700E and SF3B1WT of FLAIR isoforms with (IR) or without (spliced) retained introns. Boxplot median difference = 0.395. Source data are provided in a Source Data file. b PSI values of intron retention events identified in short read sequencing of CLL SF3B1 WT or CLL SF3B1 MT samples. Boxplot median difference = 1.69. Source data are provided in a Source Data file. c The change in PSI in significant intron retention events (corrected p < 0.1) identified in nanopore data that are more included in CLL SF3B1K700E (blue) or more included in CLL SF3B1WT (orange). Boxplot median difference = 9.32. P values for a–c are using two-sided Mann–Whitney U tests. Box-plots show median line, box limits are upper and lower quartile, and whiskers are 1.5× interquartile. d ADTRP gene isoforms, plotted 5′ to 3′. The 632 bp intron that is differentially included is boxed in purple. A differentially skipped exon is boxed in pink. e Percent usage of each isoform in each CLL sample, with colors corresponding to isoforms in d. Gray bars represent all other isoforms not plotted in d. f Top: 3′ splice-site motif of constitutively spliced introns. Bottom: 3′ splice-site motif of significant intron retention events identified from short-read sequencing (n = 67).