Abstract
Gliotoxin (GT) is a dual fungal secondary metabolite (SM). It displays pleiotropic activities and possesses medicinal properties and biocontrol abilities but, unfortunately, has toxic properties in humans. Various Trichoderma species are used as fungal biological control agents (BCAs), as a sustainable alternative for crop protection worldwide. Among them is Trichoderma virens, a GT-producing fungus. Since no information was available on the genetically coded prerequisites for the production of GT in other Trichoderma spp., genome analyses were carried out in 10 Trichoderma spp. genomes. Moreover, a real-time PCR assay setup ad hoc and high-performance liquid chromatography (HPLC) analyses were employed to understand the GT-producing biological systems in T. virens GV29-8 (TvGv29-8) and Trichoderma afroharzianum T6776 (TaT6776), two relevant biocontrol fungi. The structure of the GT biosynthesis genes (GT-BG) is polymorphic, with two distinct types associated with the ability to produce GT. GliH, a key protein for GT synthesis, is absent in most of the Trichoderma GT biosynthetic pathways, which may be the reason for their inability to produce GT. The GT-BG are expressed in TvGv29-8 as expected, while they are silent in TaT6776. Interestingly, in the GT-non-producing TaT6776, only gliA (putative GT transporter) and gtmA (putative GT S-methyltransferase) were induced by exogenous GT, underlining the ability of this strain to reduce the deleterious effect of the toxin. This ability is confirmed by growth assays and by the detection of the bis-thiomethylated form of GT catalyzed by GtmA in the culture medium supplemented with GT. To the best of our knowledge, this is the first general description of the GT biological system in different Trichoderma spp. as far as the GT-BG content and organization is concerned and a preliminary insight into their functionality.
Keywords: mycotoxin, biological control, self-protection, Aspergillus, secondary metabolism, epidithiodioxopiperazine
Introduction
Gliotoxin (GT) is a sulfur-containing fungal secondary metabolite (SM) of the class of epidithiodioxopiperazine (ETP), characterized by an internal disulfide bridge (Gardiner and Howlett, 2005; Dolan et al., 2015). Because of the presence of this bridge, GT can perform a redox cycling between the oxidized and reduced forms producing reactive oxygen species (ROS) (Gardiner and Howlett, 2005; Schrettl et al., 2010) or disturb the intracellular redox homeostasis by interacting with the thiol groups of antioxidants such as glutathione (Scharf et al., 2016).
Gliotoxin is produced by a phylogenetically diverse range of fungal species including Aspergillus fumigatus, Eurotium chevalieri, Trichoderma virens, Neosartorya pseudofischeri, and some Penicillium and Acremonium spp. This ETP can be detected in natural substrates, in the soil, in the rhizosphere, or as a contaminant of silage and other food commodities associated with the presence of fungi like A. fumigatus (Scharf et al., 2016). Remarkably, GT is produced by >96% of both environmental and clinical isolates of A. fumigatus, the opportunistic human pathogen that causes important and fatal aspergillosis, where it represents a virulence factor (Sugui et al., 2007) by means of its pleiotropic immunosuppressive activity (Konstantinovas et al., 2017).
The 13 canonical GT biosynthesis genes (GT-BG) are organized in a cluster (gli) in A. fumigatus (among others, Dolan et al., 2015), as it frequently occurs for fungal SM genes (Keller, 2019). The gli cluster includes the biosynthetic genes, the Zn(II)2-Cys(6) binuclear cluster domain transcription factor gliZ, and two self-protection genes, gliT (a GT oxidoreductase mediating the oxidation of sulfhydryl groups to form the disulfide bridge) and gliA (the GT transporter). In addition to being regulated by gliZ, both gliT, and gliA are also differentially regulated by a yet unknown regulator (Gardiner and Howlett, 2005; Schrettl et al., 2010) and by the transcription factor gipA (Schoberle et al., 2014). The environmental stimuli that induce expression of the gli cluster are still uncharacterized (Scharf et al., 2016). However, GT generally influences the expression of the biosynthetic genes in a positive feedback loop (Cramer et al., 2006). In the presence of GT, A. fumigatus produces also the bis-thiomethylated form of GT (BmGT) catalyzed by the GT S-methyltransferase GtmA. Indirectly, this enzyme negatively regulates the GT biosynthesis by disrupting the positive feedback loop, and it is not primarily involved in the self-protection mechanism (Dolan et al., 2014).
Even though GT was first discovered in T. virens (also described in literature as Gliocladium virens) (Weindling, 1934), few studies have been carried out on GT presence in this species, whose isolates were classified as “Q” strains, if they produced GT and BmGT but not gliovirin or heptelidic acid, or “P” strains, if they produced gliovirin and heptelidic acid but neither GT nor BmGT (Howell et al., 1993).
The role of GT is still debated: in A. fumigatus, it not only is a virulence factor (Sugui et al., 2007), but it also shows a protective role against oxidative stress (Owens et al., 2014); in the biological control agent T. virens, GT is thought to be linked to antagonism (Kubicek et al., 2011).
The gli cluster in T. virens is known to comprise only 8 of the 13 genes reported in A. fumigatus with two putative gliT orthologs (TRIVIDRAFT_138628 and TRIVIDRAFT_31354) and one putative gliZ ortholog (TRIVIDRAFT_191964) located elsewhere in the genome (Mukherjee et al., 2012; Vargas et al., 2014). Besides these, to the best of our knowledge, the only other reports on GT-BG in Trichoderma spp. refer to Trichoderma reesei where either a truncated GT cluster with only 8 of the 13 gli genes (Mukherjee et al., 2012) or an ETP gene cluster of 12 genes (Patron et al., 2007) were described; importantly, both reports agreed on the absence of GT production by the T. reesei strains.
Trichoderma isolates are among the most frequently used microorganisms as active principles of commercial biofertilizers and biopesticides, yet nine species among which T. longibrachiatum, T. atroviride, T. harzianum, T. reesei, and T. viride are infrequent but emergent opportunistic human pathogens (Druzhinina et al., 2008; Sandoval-Denis et al., 2014; Sautour et al., 2018). Although biopesticides derived from naturally occurring microorganisms are generally considered to provide an environmentally benign pest control option, they may not be entirely free of hazards and consequently require an accurate risk assessment (Konstantinovas et al., 2017; Bulgari et al., 2019).
Although GT represents a virulence factor for A. fumigatus, few studies aimed to monitor the presence of GT or the gli biosynthesis genes (GT-BG) in Trichoderma spp.
In the present study, the GT-BG structure in six representative Trichoderma species was obtained, taking advantage of the fact that several genome sequences of Trichoderma species are now available, providing the opportunity to rapidly screen fungal genomes for the presence of the gli cluster and compare its organization between different species. Moreover, the GT biological system was characterized by high-performance liquid chromatography (HPLC) analyses in the agriculturally relevant strains T. virens Gv29-8 and T. afroharzianum T6776 and a useful real-time reverse transcription PCR (RT-PCR) assay was developed ad hoc to follow the expression of their gli genes encoding GliP, GliT, GliA, GliH, GipA, and GtmA in GT permissive conditions.
Materials and Methods
Strains and Growth Conditions
The experimental studies were carried out on the biocontrol agent T. virens Gv29-8, a GT-producing strain, and on the plant growth promoter T. harzianum T6776 (Fiorini et al., 2016), recently reclassified as T. afroharzianum T6776 (Kubicek et al., 2019). The fungal strains were routinely cultivated on potato dextrose agar (PDA) or potato dextrose broth (PDB) as specified.
Bioinformatic Analyses of Gliotoxin Biosynthesis Genes
The fungal strains, whose genome sequences were downloaded from the National Center for Biotechnology (NCBI) for the in silico study, are presented in Table 1 with their related abbreviations and other information. A. fumigatus Af293 was used as a reference strain and its annotated genome as query. Different basic local alignment search tool (BLAST) analyses (blastn, tblastn, blastp, blastx) were performed using the Af293 gli cluster gene and protein sequences as a query to further assess content and arrangements of genes in TvGv29-8. Successively, the TvGv29-8 gli genes and protein sequences were also used as references to find the putative orthologous gli genes among Trichoderma spp. For the gliH search, the AfgliH described by Schrettl et al. (2010) was used instead of AFUA_5G10320, a gene annotated as gliH in the NCBI database. The highest scoring and covering hit for each gene was verified by the bidirectional best hits (BBH) approach against Af293 and the other Trichoderma genomes and eventually chosen, identified, and localized on the Trichoderma spp. genomes. Each gene orthology was then verified in the OrthoDB catalog of eukaryotic orthologs provided as an option in NCBI. To clearly distinguish between orthologous genes in A. fumigatus, T. virens, T. afroharzianum, T. harzianum, T. reesei, T. gamsii, and T. longibrachiatum, gene designations are preceded, respectively, by the prefix Afgli, Tvgli, Tagli, Thgli, Trgli, Tggli, and Tlgli. The Afgli genes and putative GT-BG of the analyzed Trichoderma strains are listed in Supplementary Table S1.
TABLE 1.
Fungal strains used or whose genomes were used in this study.
| Organism | Strain | Collection | NCBI Bioproject no. | Designation in this study |
| Aspergillus fumigatus | Af293 | NCPF 7367, CBS 101355 | PRJNA131 | Af293 |
| Trichoderma virens | Gv29-8 | ATCC MYA-4894 | PRJNA19983 | TvGv29-8 |
| FT-333 | National Chiayi University, Taiwan | PRJNA268050 | TvFT333 | |
| Trichoderma afro-harzianum | T6776 | University of Pisa, Italy | PRJNA252551 | TaT6776 |
| Trichoderma harzianum | B97 | Biovitis, France | PRJNA357189 | ThB97 |
| TR274 | Universidade Fed. de Goias, Goiânia, Brazil | PRJNA397414 | ThTR274 | |
| CBS 226.95 | CBS 226.95 | PRJNA207867 | Th226.95 | |
| Trichoderma reesei | RUT-C30 | ATCC 56765 | PRJNA207855 | TrRUTC30 |
| QM6a | ATCC 13631 | PRJNA15571 | TrQM6a | |
| Trichoderma gamsii | T6085 | University of Pisa, Italy | PRJNA252048 | TgT6085 |
| Trichoderma longibrachiatum | SMF2 | Shandong University, Jinan, China | PRJNA175761 | TlSMF2 |
Selection of Candidate Genes and Primers Design
To verify the correct identification and functionality of the gli clusters in TvGv29-8 and TaT6776, candidate reference genes for gene expression analyses were shortlisted. In detail, gliP, coding the key GT biosynthetic bimodular non-ribosomal peptide synthetase; gliH, unknown function; gliT and gliA, self-protection genes; gliZ and gipA, transcription factors; and gtmA, a gene belonging to a negative regulatory system, were chosen as the main representative genes for GT biosynthesis, self-protection, and regulation, respectively. Calmodulin (TCal) and secretion-associated Ras-related protein (TSar) genes were used as housekeeping genes for gene expression normalization. Real-time RT-PCR protocols were developed for all these genes. Each primer was designed in an intron flanking region. Specific primer pairs were designed using Primer3Plus. The gene names, amplification efficiency, correlation coefficient, amplicon length, and amplicon melting temperature are reported in Supplementary Table S2. The amplification efficiency of quantitative PCR (qPCR) was calculated only for the genes expressed in the absence and the presence of exogenous GT and ranged from 90 to 99.8%, and the R2 of the standard curve was 0.997 ± 0.001. Agarose gel electrophoresis and melting curve analyses (data not shown) showed a single temperature for each primer pair that was distinctly different from the non-specific genomic peak, confirming the specificity of the amplification.
RNA Extraction and Real-Time PCR
TvGv29-8 and TaT6776 conidia were inoculated in PDB at a final concentration of 107 conidia/ml and incubated at 23–25°C under illumination of 16 h light/8 h dark cycles, using daylight tubes 24 W/m2, 9,000 lx. After 27 h, fungal RNA was isolated and purified using the PureLink RNA purification kit (Ambion, Thermo Scientific). To avoid genomic DNA (gDNA) contaminations, the RNAs were treated with DNase I (Ambion, Thermo Scientific), and complementary DNA (cDNA) synthesis from messenger RNA (mRNA) (200 ng) was performed using cDNA first-strand synthesis kit (Thermo Fisher) with hexamer primer according to manufacturer’s instruction.
Reactions were performed using the Real-Time PCR PowerUp SYBR Green Master Mix kit (Applied Biosystems) in the ViiA7 Real-Time PCR system (Applied Biosystems). Each RT-PCR reaction was performed in three biological and technical replicates and a no-template control. To determine the specificity of the primer pairs used in the current study, 1.5% agarose gel electrophoresis and melt curve analyses were performed (Supplementary Table S2). To determine the PCR efficiency of each primer pair, a standard curve was generated using linear regression, and the Cq slope was calculated based on the Cq values for all dilutions (5 points, 10-fold dilutions from 50 to 50,000).
Induction With Exogenous Gliotoxin
TvGv29-8 and TaT6776 were inoculated in PDB by spore suspension at a final concentration of 107 conidia/ml and incubated at 23–25°C under illumination of 16 h light/8 h dark cycles, using daylight tubes 24 W/m2, 9,000 lx. After 24 h, exogenous GT (WVR International) or methanol (MeOH, solvent control) was added to the media at 5 μg/ml (final concentration) and incubated for 3 h under the same conditions. Mycelia were harvested through Miracloth filtration, washed with distilled water, and immediately frozen in liquid nitrogen and processed for gene expression analyses as described above. The culture filtrates were stored at −20°C until GT identification and quantification by HPLC analysis.
GT and BmGT Production in Liquid Media
Conidia (1 × 106 conidia) of TvGv29-8, TaT6776, and TrRUTC30 (as negative control) were inoculated into 25 ml Weindling modified medium (Wilhite and Straney, 1996) and cultured on a rotary shaker (150 rpm) at 25°C of 16 h light/8 h dark cycles, using daylight tubes 24 W/m2, 9,000 lx. To investigate the time course of GT production, aliquots of the culture filtrates were taken at incubation times 24, 48, 72, 96, and 162 h and analyzed by HPLC. Three replicates were performed for each fungus. For BmGT quantification, samples were taken at 27 h incubation time.
Extraction and Estimation of Gliotoxin by HPLC
Culture filtrate aliquots were diluted with MilliQ water (1:1) and extracted three times with half volume of chloroform while shaking (150 rpm) for 10 min. The concentrated chloroform extracts were dried over sodium sulfate, and the solvent was removed by Rotavapor (37°C, vacuum); the residue was dissolved in 200 μl methanol. Only the samples induced with exogenous GT were furtherly diluted 1:10. All the samples were then subjected to HPLC analysis.
Ten microliters of each biological sample was analyzed in triplicate by reverse-phase HPLC with UV detection (RP-HPLC-UV) using a DionexTM UltiMateTM 3000 (Thermo Fisher Scientific) equipped with an LPG-3400SD quaternary analytical pump, a WPS-3000SL analytical autosampler, a VWD-3100 UV–visible detector, a TCC-3000SD thermostatted column compartment, and an AFC-3000 automatic fraction collector. Chromatographic separation was performed using a Waters XSelect CSH C18 column (150 mm × 2.1 mm ID; particle size, 3.5 μm). The specific liquid chromatographic (LC) parameters were as follows: mobile phase (A) water, TFA (99.95:0.05 v/v); (B) acetonitrile, the mobile phase flow rate was 0.3 ml/min; gradient program, 10% B for 2 min, from 10 to 90% B in 18 min, 90% B for 1 min and then from 90 to 10% B in 1 min and re-equilibration to 10% B for 3 min. All analyses were performed at 30°C. The detection wavelength (λ) was set at 254 nm.
As reference standard GT (VWR International) and BmGT (Sigma-Aldrich Co.) were used. For GT quantitation, a calibration curve was generated using five different dilutions of the GT reference standard (550.00, 275.00, 55.00, 27.50, and 5.50 μg/ml) and similarly for BmGT quantitation, using five different dilutions of the BmGT reference standard (50.00, 25.00, 10.00, 5.00, 1.00 μg/ml). Ten microliters of each standard dilution was analyzed by RP-HPLC-UV as reported above. The calibration curve was plotted using the area under peak versus GT or BmGT standard reference concentrations (μg/ml) (Supplementary Figures S1A,B, respectively). Biological sample concentrations were calculated by determining their area under peak and comparing them to the area of the calibration curve.
Trichoderma Multiwells Growth Assay With Exogenous GT
TvGv29-8 and TaT6776 conidia at a final concentration of 106 conidia/ml were inoculated in PDB containing different concentrations of GT or MeOH (0, 5, 15, 30, 50 μg/ml) in 96-well plates. The optical density (620 nm) of the liquid cultures was read at least three times a day at regular intervals, using an ELISA plate reader.
Results
In silico Identification of the GT Biosynthesis Genes and Related Genes in Trichoderma virens
Three new putative orthologs – TvgliJ, TvgliA, and TvgliH encoding a dipeptidase, a transporter, and an unknown function protein, respectively – were identified in TvGv29-8, at other loci, outside of the previously described gli cluster on scaffold 47. The relative position and characteristics of the Tvgli genes are shown in Table 2 and Supplementary Table S1. In detail, TvgliJ, located on scaffold 51, encodes a protein with a 52.81% sequence identity to AfGliJ; TvgliA, situated on scaffold 4, codes for TvGliA with 45.13% sequence identity to AfGliA; and TvgliH, on scaffold 10, encodes TvGliH with 54.40% sequence identity to AfGliH. Interestingly, of the TvgliT orthologs previously described, TRIVIDRAFT_138628 was found beside TvgliH, while TRIVIDRAFT_31354 was located on the large scaffold 4 where TvgliA is also present. In addition, two related genes, TvgtmA, the GT thiomethyltransferase, and TvgipA, a C2H2 transcription factor, were identified on TRIVI scaffolds 77 and 5, respectively.
TABLE 2.
Sequence similarity analyses of the gli gene orthologs in T. virens, T. afroharzianum, T. reesei, and T. longibrachiatum.
|
T. virens Gv 29-8 |
T. afroharzianum T6776 |
T. reesei RUT-C30 |
T. reesei QM6a |
T. longibrachiatum SMF2 |
|||||||||||||||
| Gene | Query | E value | Ident | Gene | Query | E value | Ident | Gene | Query | E value | Ident | Gene | Query | E value | Ident | Gene | Query | E value | Ident |
| TvgliZ | 26% | 1e-20 | 53% | TagliZ | – | – | – | TrgliZ | – | – | – | TrgliZ | – | – | – | TlgliZ | – | – | – |
| TvgliI | 96% | 1e-155 | 52% | TagliI | 96% | 8e-91 | 35% | TrgliI | 96% | 4e-108 | 39% | TrgliI | 97% | 1e-108 | 40% | TlgliI | 96% | 1e-88 | 34% |
| TvgliJ | 99% | 2e-149 | 53% | TagliJ | 92% | 2e-133 | 50% | TrgliJ | 95% | 3e-144 | 55% | TrgliJ | 95% | 3e-144 | 55% | TlgliJ | 92% | 2e-122 | 47% |
| TvgliP | 95% | 0.0 | 44% | TagliP | 95% | 0.0 | 35% | TrgliP | 98% | 0.0 | 40% | TrgliP | 98% | 0.0 | 40% | TlgliP | 95% | 0.0 | 38% |
| TvgliC | 90% | 8e-157 | 48% | TagliC | 89% | 9e-131 | 43% | TrgliC | 96% | 1e-155 | 46% | TrgliC | 89% | 9e-155 | 49% | TlgliC | 87% | 9e-151 | 48% |
| TvgliM | 96% | 5e-174 | 56% | TagliM | 87% | 1e-119 | 45% | TrgliM | 85% | 8e-122 | 47% | TrgliM | 85% | 8e-122 | 47% | TlgliM | 77% | 5e-120 | 51% |
| TvgliG | 95% | 5e-113 | 68% | TagliG | 78% | 8e-37 | 38% | TrgliG | 82% | 8e-55 | 43% | TrgliG | 82% | 8e-55 | 43% | TlgliG | 78% | 2e-41 | 41% |
| TvgliK | 89% | 7e-83 | 47% | TagliK | 94% | 2e-53 | 38% | TrgliK | 94% | 6e-66 | 40% | TrgliK | 94% | 6e-66 | 40% | TlgliK | 95% | 6e-44 | 44% |
| TvgliA | 98% | 1e-160 | 45% | TagliA* | 26% | 0.39 | 40% | TrgliA* | 16% | 0.90 | 23% | TrgliA* | 16% | 0.82 | 23% | TlgliA* | 19% | 1.4 | 23% |
| TvgliN | 98% | 4e-123 | 58% | TagliN | 97% | 2e-32 | 26% | TrgliN | 97% | 4e-32 | 27% | TrgliN | 97% | 4e-32 | 27% | TlgliN | 96% | 2e-27 | 26% |
| TvgliF | 99% | 0.0 | 61% | TagliF | 39% | 7e-18 | 27% | TrgliF | 65% | 6e-20 | 23% | TrgliF | 88% | 1e-58 | 27% | TlgliF | 44% | 1e-10 | 27% |
| TagliF° | 34% | 1e-11 | 24% | TrgliF° | 34% | 1e-09 | 23% | TrgliF° | 34% | 2e-09 | 23% | ||||||||
| TvgliT* | 86% | 1e-115 | 58% | TagliT | 93% | 8e-42 | 33% | TrgliT | 92% | 1e-53 | 36% | TrgliT | 92% | 1e-53 | 36% | TlgliT | 91% | 2e-44 | 33% |
| TvgliT** | 87% | 3e-107 | 52% | ||||||||||||||||
| TvgliH | 96% | 5e-68 | 54% | TagliH | – | – | – | TrgliH | – | – | – | TrgliH | – | – | – | TlgliH | – | – | – |
| TvgipA | 60% | 5e-44 | 51% | TagipA | 97% | 4e-48 | 36% | TrgipA | 48% | 4e-44 | 51% | TrgipA | 48% | 4e-44 | 51% | TlgipA | 35% | 5e-30 | 41% |
| TvgtmA | 87% | 7e-43 | 32% | TagtmA | 87% | 5e-41 | 31% | TrgtmA | 26% | 3e-04 | 34% | TrgtmA | 92% | 8e-41 | 30% | TlgtmA | 90% | 8–16 | 31% |
The results are indicated as Query coverage (Query), BLAST E value (E value), and identity percentage (Ident) of deduced protein sequences with A. fumigatus gli proteins as references. gliA*, ABC transporters; putative orthologs present in T. afroharzianum, T. reesei, and T. longibrachiatum; gliT*, putative ortholog corresponding to TRIVIDRAFT_138628; gliT**, putative ortholog corresponding to TRIVIDRAFT_31354; gliF°, putative 2° ortholog present in T. afroharzianum and T. reesi.; – not detected.
Overall, TvGv29-8 and Af293 showed a gli-gene-related protein sequence identity between 44 and 68% (Table 2). Thus, in addition to the cluster of eight gli genes, a duplet of two gli genes, two single gli genes, situated in different locations in the genome, and two related genes were identified (Figure 1).
FIGURE 1.
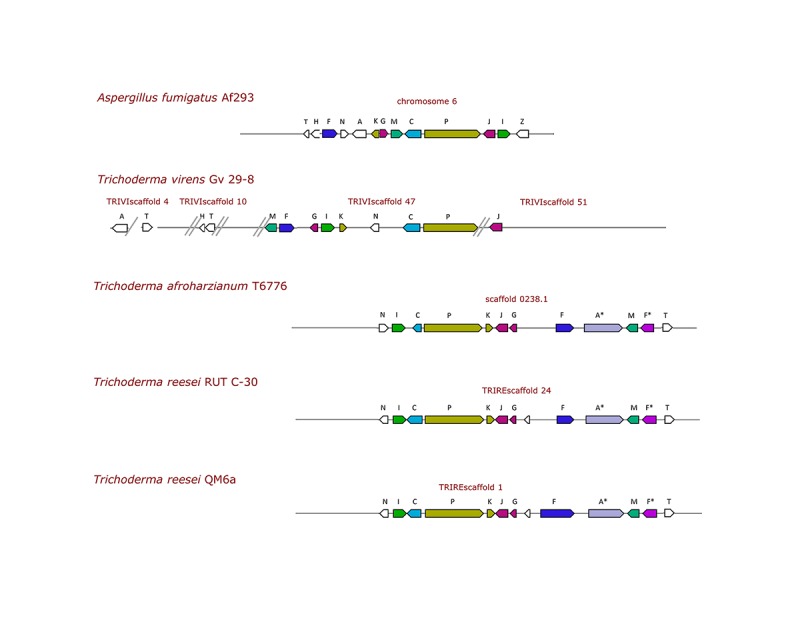
Schematic view of the gliotoxin biosynthesis genes (GT-BG) in A. fumigatus and representative Trichoderma spp. strains. Arrows represent the genes, and their size is proportional to the length of the corresponding genomic region. The GT-BG are gliZ (Z), zinc finger transcription factor; gliI (I), 1-aminocyclopropane-1-carboxylate synthase; gliJ (J), a dipeptidase; gliP (P), a two-module non-ribosomal peptide synthetase (NRPS); gliC (C), gliF (F), two cytochrome P450 monooxygenases; gliF* (F*), second putative ortholog; gliM (M), O-methyltransferase; gliG (G), a glutathione-S-transferase; gliK (K), a cyclotransferase (GGCT); gliA (A) or gliA* (A*), respectively, major facilitator superfamily (MFS) or ATP-binding cassette (ABC) transporter; gliN (N), a methyltransferase; gliT (T), a GT oxidase; gliH (H), coding a conserved hypothetical protein. The scaffolds containing the GT-BG are separated by double lines at their extremities; a single line indicates that the genes are present in the same scaffold but not clustered.
When a similar to GT-BG and related genes search was performed in TvFT-333, using the TvGv29-8 gene sequences, a striking nucleotide identity (100%) (Supplementary Table S3A) was found with the corresponding 13 Tvgli genes (Supplementary Table S4). In addition, TvFT-333 showed an identical GT-BG architecture with a cluster of the same eight gli genes with the same order and orientation (on scaffolds 0430–0433) and two single gli genes, gliA and gliJ, situated respectively, in scaffolds 0040 and 0450. Differently, of the duplet of two gli genes, only TvgliT was present on scaffold 0048, while TvgliH was absent from the TvFT-333 genome. This result was confirmed also by blasting the TvGliH and AfGliH sequences.
To verify the distribution of the GT-BG in different loci, the sequences of the flanking regions of the gli cluster, the duplet and the single genes were compared in the two T. virens strains. Actually, in TvGv29-8, the gli cluster, the gliH-gliT duplet, and one end of gliJ are without flanking sequences (Figure 1). Differently, the sequences flanking gliA and the other end of gliJ confirmed their position outside of the gli cluster.
The third T. virens genome present in NCBI belongs to the “P” strain, IMI 30406, which does not contain any gli genes (Sherkhane et al., 2017); thus, it was not included in this study.
In silico Identification of the GT Biosynthesis Genes and Related Genes in Trichoderma spp.
Putative gli clusters were successfully identified in seven strains of four different species: TaT6776, ThB97, ThTR274, Th226.95, TrRUTC30, TrQM6a, and TlSMF2 (Table 2 and Supplementary Tables S1,S4). GT-BG of some representative strains are depicted in Figure 1, where the multiple genes shared between Trichoderma species are shown. Interestingly, the gli cluster was not present in the TgT6085 genome. The organization of the GT-BG in these Trichoderma species differed considerably from T. virens, that is, the GT-BG were present in a single cluster with a very similar architecture: length ranging from 33,140 to 35,284 bp, presence of 11 gli genes, and absence of gliH and gliZ (Supplementary Table S5). Notably, the T. reesei GT-BG were located in a single cluster with 11 out of the 12 genes reported by Patron et al. (2007).
Table 2 and Supplementary Table S4 show the highest identities found in protein sequences using BLASTP search. The predicted products of the gli genes were similar to those found in A. fumigatus showing amino acid identities ranging from 23 to 82%.
Moreover, a very high (99.97%) intraspecies nucleotide identity of the gli clusters was found in T. harzianum and T. reesei, while interspecies identity varied from 70% between Tagli/Trgli and Tagli/Tlgli, to 88% between Tagli/Thgli. In these species, one major facilitator superfamily (MFS) transporter highly similar to AfgliA was located outside of the clusters, while an ATP-binding cassette (ABC) transporter was located in the gli clusters and coding for proteins with a low amino acid sequence identity to AfGliA and TvGliA (Supplementary Table S3B). These ABC transporters (gliA∗) showed very high interspecies identity (70–88%). The MFS and ABC transporter genes were found to belong to two different orthology groups. Lastly, gliZ relics were found in a genomic position located close to gliG and a second putative gliF, which resulted not to be an AfgliF ortholog. All the GT-BG identified in these species were indicated as orthologs of the corresponding Afgli genes by OrthoDB.
GtmA and gipA were present in all the Trichoderma species tested (Table 2 and Supplementary Table S4). Notably, orthologs of TvgtmA (TGAM01_v208885) and of AfgipA and TvgipA (TGAM01_v202426) were also found in TgT6085, where the GT-BP was absent.
Gene Expression
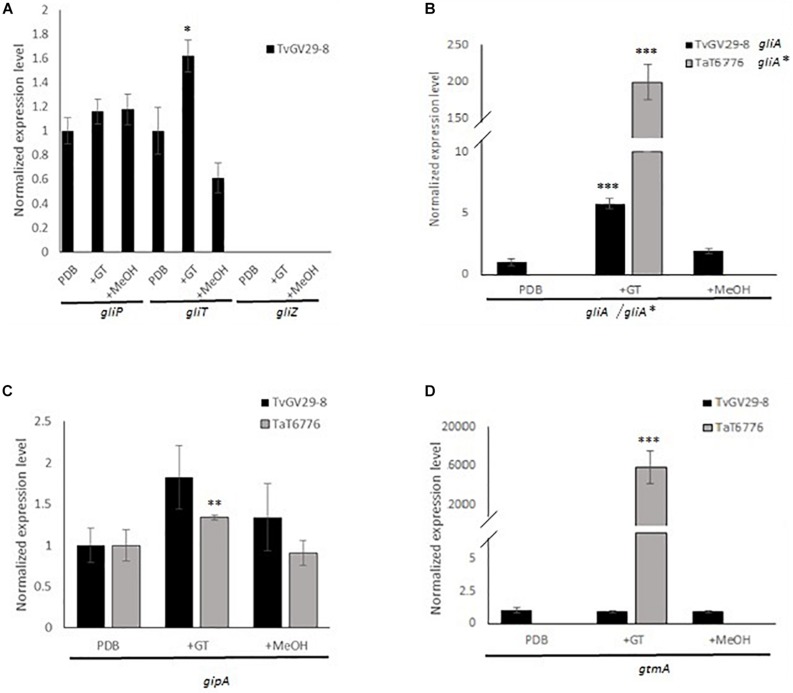
The expression levels of gliP (non-ribosomal peptide synthetase), gliT (putative oxidoreductase), gliA (putative gliotoxin transporter), gliZ (putative Zn2Cys6 transcription factor), gliH (hypothetical protein), gipA (putative C2H2 transcription factor), and gtmA (putative GT bis-thiomethyltransferase) were analyzed (Figure 2). The results were quite different between the two Trichoderma strains. In the GT-producing TvGv29-8, TvgliP, the key gene in GT synthesis, as well as TvgliA, TvgipA, and TvgtmA were expressed. Among the previously identified gliT orthologs, only gliT (TRIVIDRAFT_138628) was expressed during GT production, establishing it as the putative oxidoreductase TvgliT. Otherwise, TvgliZ is still to be identified since neither the previously indicated ortholog nor the other hits revealed by BLAST analyses (data not shown) were expressed. Interestingly, gliH that was retrieved only in the TvGv29-8 genome was expressed in GT permissive conditions (data not shown). In TaT6776, only gipA was transcribed. The TvgliH primers were also tested in TaT6776, but no amplifications were observed.
FIGURE 2.
Compared gene expression analyses in TvGv29-8 and TaT6776 grown in potato dextrose broth (PDB), in PDB amended with 5 μg/ml of gliotoxin (+GT), or amended with methanol (+MeOH). (A) gliP, gliT, and gliZ in TvGv29-8, no transcriptions were detected in TaT6776; (B) gliA/gliA* [major facilitator superfamily (MFS)/ATP-binding cassette (ABC) transporter]; (C) gipA; (D) gtmA. Each real-time PCR (RT-PCR) reaction was performed in three biological and technical replicates. For statistical analysis, ANOVA test was applied with significance defined as a p value (*p ≤ 0.02; **p ≤ 0.001, ***p ≤ 0.0001).
Expression of GT Biosynthesis Genes in the Presence of Exogenous GT
In A. fumigatus, AfgliP, AfgliT, AfgliA, and AfgipA are involved both in GT production and self-protection against GT toxicity (Schrettl et al., 2010; Dolan et al., 2015), while AfgtmA indirectly and negatively regulates the production of GT (Dolan et al., 2017), which is known to be involved in a positive regulation of its own synthesis (Cramer et al., 2006). To evaluate the role of exogenous GT on gliP, gliT, gliA, gipA, and gtmA expression, RT-qPCR results were compared between TvGv29-8 and TaT6776 grown in different conditions (PDB, 5 μg/ml GT, and methanol), and the significance was evaluated by ANOVA (Figure 2).
In detail, in TvGv29-8, TvgliP and TvgtmA expression levels were not significantly different in all the tested conditions. In contrast, TvgliT and TvgliA genes were upregulated by 1.6- and 6-fold, respectively, in the presence of exogenous GT. The expression levels of the transcription factor gipA were increased in both TvGv29-8 and TaT6776 in the presence of exogenous GT, but this effect was statistically significant only in TaT6776 (p ≤ 0.001). Strikingly, when the fungus TaT6776 was exposed to exogenous GT, the expression of the TagliA∗ and TagtmA genes was detectable and measured by 200- and 16,000-fold, respectively, compared to samples not exposed to GT (p ≤ 0.0001). The expression of the tested genes in the solvent control was comparable to that observed without exogenous GT.
T. afroharzianum Is a GT-Non-producing Strain
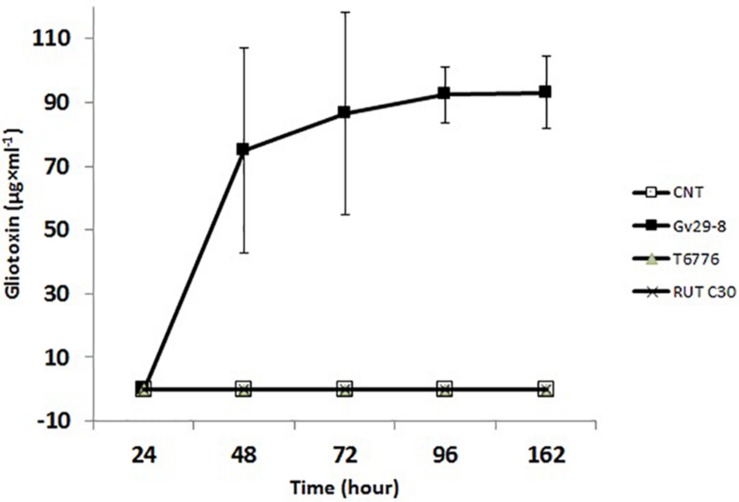
TvGv29-8 is a well-known GT-producing strain, which was confirmed in this study by HPLC analyses. The GT and BmGT concentrations referred to the level present in the original culture media. GT production began at 24 h after inoculation in Weindling medium, and it rapidly increased to a concentration of 74.9 ± 32.3 μg/ml at 48 h (Figure 3). Subsequently, the GT secreted was slowly increased to 93.1 ± 11.3 μg/ml at 162 h.
FIGURE 3.
Gliotoxin time course production in TvGv29, TaT6776, and TrRUTC30, grown in Weindling medium up to 162 h, detected by high-performance liquid chromatography (HPLC). Data points represent the average ± standard deviation of biological replicate cultures (n = 3).
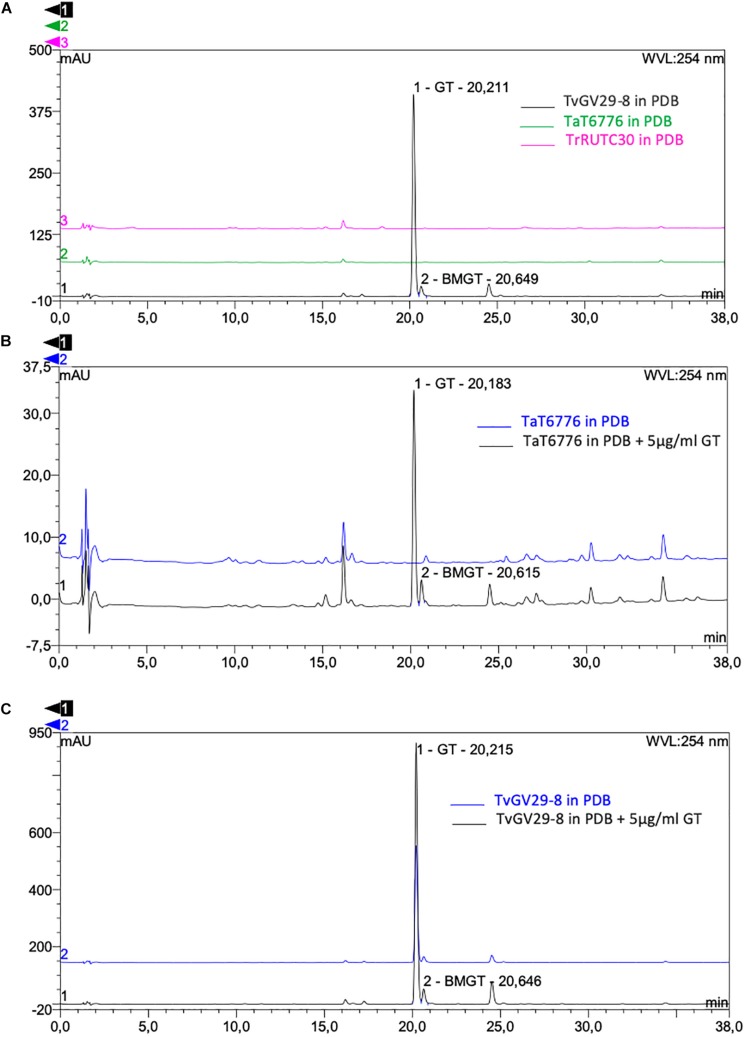
No GT- and BmGT-related peaks were present for TaT6776 (Figure 4A), demonstrating that it does not produce GT, despite the presence of the gli cluster in its genome. No information was available on GT production by TaT6776 before this study. As expected, comparable HPLC results were found for TrRUTC30, confirming that it does not produce GT (Figure 4A).
FIGURE 4.
High-performance liquid chromatography (HPLC) detection of gliotoxin (GT) and bis(methyl)gliotoxin (BmGT) in fungal culture filtrates. (A) Comparison between TvGv29-8 (black), TaT6776 (green), and TrRUTC30 (magenta). The chromatograms show the presence of GT and BmGT only in TvGv29-8 sample (black). HPLC detection of gliotoxin (GT) and bis(methyl)gliotoxin (BmGT) in culture filtrates after 27 h incubation time of (B) TaT6776 and (C) TvGv29-8, grown in potato dextrose broth (PDB) (blue) and in PDB amended with 5 μg/ml of GT (dark gray). HPLC analysis was performed in four technical replicates.
Exogenous Gliotoxin Induces Bis(Methylthio)gliotoxin Production in Trichoderma spp.
The production of GT and BmGT in TvGv29-8 and in TaT6776 was analyzed further after adding exogenous GT to their culture media. In this case, both GT and BmGT were present also in the culture filtrates of TaT6776 (Figure 4B). The amount of recovered GT (4.98 ± 0.9 μg/ml) corresponded to that added to the medium, while 0.19 ± 0.07 μg/ml of BmGT was produced by TaT6776.
In TvGv29-8, the increased amount of synthesized GT, from 67.09 ± 28.6 μg/ml without the addition of exogenous GT to 93.4 ± 37 μg/ml with the addition of exogenous GT, was not statistically significant (Figure 4C). Similarly, in TvGv29-8, BmGT production was observed at 27 h after inoculation at a concentration of 0.98 ± 0.4 μg/ml and 1.57 ± 0.8 μg/ml (Figure 4A), respectively, without and with the addition of exogenous GT.
T. afroharzianum Is Able to Grow in the Presence of Gliotoxin
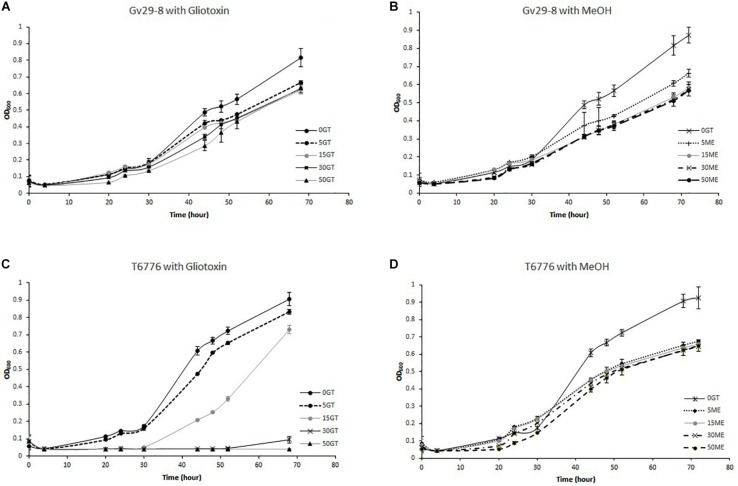
The ability of TvGv29-8 and TaT6776 to grow at different GT concentrations in the culture medium was verified in a multiwell assay (Figures 5A,C). The fungi differed in their sensitivity to exogenous GT. TvGv29-8 growth was only slightly affected by any GT concentration up to 50 μg/ml. In contrast, the growth of TaT6776 was completely suppressed at 50 μg/ml GT and significantly inhibited (p = 0.005) at 30 μg/ml GT. Nonetheless, TaT6776 did not show a dramatic sensitivity to exogenous GT, since the inhibition of its growth at 15 μg/ml GT was small (19%), although statistically significant (p = 0.003), compared to its growth without GT. Similarly, its growth was inhibited by 25% at 30 μg/ml GT after 96 h of incubation (data not shown). Interestingly, the presence of methanol in the media did not appear to cause a relative reduction in the growth of both TvGv29-8 and TaT6776 even at the volumes used at the highest tested GT concentrations, 30 and 50 μg/ml (Figures 5B,D).
FIGURE 5.
Growth of (A) TvGv29-8 and (C) TaT6776 at different concentrations of exogenous gliotoxin (0–5–15–30–50 μg/ml). Growth of (B) TvGv29-8 and (D) TaT6776 at different volumes of methanol corresponding to those present in the different gliotoxin (GT) concentrations. Data points represent the average ± standard deviation of biological replicate cultures (n = 3).
Discussion
Fungal SMs are defined as molecules not strictly necessary for life but critical for the lifestyle of their producers (Keller, 2019), largely conferring a better response to ecological pressures and regulated by the different environments (Bayram et al., 2008; Reverberi et al., 2010).
In T. virens, frequently used as biological control agent (BCA), GT is associated with its ability to control plant pathogens (Scharf et al., 2016). However, SMs are also a health concern because they represent often conserved trait associated to virulence of pathogenic fungi (Bergmann et al., 2007; Wisecaver and Rokas, 2015). An example is A. fumigatus where GT is considered a virulence factor (Sugui et al., 2007), and consequently, GT biosynthesis, functionality, and role have been studied in depth.
Since GT shows beneficial and deleterious functions, depending on the context, there is a need for extensive knowledge of the ability of agricultural BCAs to produce GT. To the best of our knowledge, no studies have examined the genetic potential for GT production in T. afroharzianum, T. harzianum, T. longibrachiatum, and T. gamsii. Moreover, the identification of the GT-BG of T. virens and T. reesei was extended and combined with preliminary insights into their functionality.
In this work, the Trichoderma GT-BG were searched in 6 out of the 19 species present in NCBI, for a total of 10 analyzed genomes.
Gliotoxin biosynthesis genes are quite common in Trichoderma spp. They are present in five species – T. virens, T. harzianum, T. afroharzianum, T. reesei, and T. longibrachiatum – and absent only in T. gamsii, among those examined in this study.
A clear polymorphism of the GT-BG structure was revealed by in silico analyses, and two distinct types of organization were identified. The first type was the characteristic of T. virens Q strains where the identification of three new putative Tvgli genes extended the GT-BG to 12. These genes were located at four distinct loci, and the gli cluster was separated from other essential genes (gliA; gliH-gliT duplet; gliJ). To assess if the GT-BG separation could be due to an incorrect genome assembly, the comparison of the two T. virens genomes present in NCBI was performed. Unfortunately, the absence of flanking sequences of the gli cluster and the gliH–gliT duplet in TvGv29-8 and the shortness of the contig length in TvFT-333 did not allow to ascertain their real position. Nonetheless, gliJ and gliA were found to be separated from the gli cluster in both strains. These findings, together with the same shared overall GT-BG structure, support the location at different loci of the GT-BG in T. virens.
In fungi, genes involved in SM biosynthesis are generally clustered in the genome, are coordinately regulated, and typically encode biosynthetic enzymes, one or more transcription factors, and a transporter (Nützmann et al., 2018). Nonetheless, they maintain their functionality even if located at different loci in the genome as in the trichothecene biosynthetic pathway in Fusarium spp. (Proctor et al., 2009). The second polymorphic type was characterized by a cluster in a single locus as described for A. fumigatus; it contained 11 gli genes and was characterized by the absence of gliH and by the presence of a gene coding a different class of transporter. In both GT-BG structures, the transcription factor gliZ was not identified.
These findings raise the questions: did gliA, gliT, gliH, and gliJ originate from the core cluster and were subsequently relocated, or did these genes originate outside the cluster and enter it? Why is gliH commonly absent in the Trichoderma genome, but so far has been found only in TvGv29-8? Does gliZ exist in Trichoderma spp.? Additional experiments are warranted to answer these questions, but were beyond the scope of this study.
The GT-BG structure polymorphism appears to be more associated with GT production than with fungal taxonomic status. In fact, while the first type was present only in T. virens in the GT-producing TvGv29-8, the second type was present in four species, among which two were proven unable to produce GT. The second type was also more conserved and was characterized by a high level of DNA sequence homology and by the same number, order, and orientation of the gli genes. Of the two T. virens genomes examined here, TvGv29-8 and TvFT-333, although they share an extremely high sequence identity of their GT-BG, differ in the presence/absence of TvgliH. Of course, these results, derived from a limited number of strains of these species, should be considered preliminary so far. However, they shed a light on a relevant fungal SM system.
Here, for the first time, the functionality of the GT-BG of Trichoderma spp. was analyzed using RT-qPCR developed ad hoc in this study. This circumscribed comparison of the GT-BG between GT-producing and GT-non-producing strains highlighted their active transcription only in TvGv29-8. The gene expression analyses combined with HPLC revealed that the key genes for GT biosynthesis are not expressed in TaT6776, which consequently fails to produce GT in these experimental conditions. The only gene expressed in default conditions in TaT6776 was gipA, a transcription factor, possibly involved in other biosynthetic pathways. Because metabolite production is influenced by environmental stimuli, these findings, obtained under laboratory culture conditions (i.e., on Weindling and PDB media), do not exclude the possibility of GT production on other media and under different environmental conditions. However, on the basis of the GT-BG content, the majority of the analyzed Trichoderma spp. strains appears unable to produce GT. A tentative explanation of this behavior could dwell on (i) the absence of gliH, (ii) the lack of expression of gliT, and (iii) possible mutation or deletion of the regulatory gene gliZ. In A. fumigatus, gliH and gliT are considered to be linked to biosynthetic genes, and their mutants ΔgliH26933 and ΔgliT26933 are unable to support production of GT (Schrettl et al., 2010). In Trichoderma spp., gliH is present only in the GT-producing TvGv29-8 strain and, furthermore, is located in duplet with the expressed TvgliT. On the basis of this work, it is possible to hypothesize that the lack of gliH and/or the absence of transcription of gliT could determine the inability of Trichoderma spp. to produce GT. The lack of gliH in TaT6776 genome is supported by the absence of its amplification by PCR in this fungus; on the other hand, the gliH absence possibly due to genome sequencing incompleteness cannot be ruled out owing to the impossibility to design a proper PCR protocol. Conversely, gliH was expressed in TvGv29-8 in GT permissive conditions.
As Trichoderma spp. have a comparable genetic machinery to A. fumigatus, a similar function of the GT biological system is hypothesized. A. fumigatus evades the harmful effects of GT by the combined activity of GliT and GliA in the so-called self-protection mechanism (Wang et al., 2014; Dolan et al., 2015). Upon exposure of TvGv29-8 to exogenous GT, TvgliT and TvgliA were overexpressed, suggesting a possible role of the encoded proteins in the protection from the GT deleterious effects. Strikingly, exposure of the GT-non-producing TaT6776 strain to exogenous GT strongly induced TagliA∗ expression but not that of TagliT. In A. fumigatus, the gli cluster is coordinately regulated by AfgliZ, but some gli genes show also different regulation. AfgliT, for example, is regulated either coordinately or in an independent manner by AfgliZ and a still unknown regulator (Schrettl et al., 2010), and AfgliA is under the interdependent control of AfgliZ and AfgipA (Schoberle et al., 2014). The expression of TagliA∗ and the contextual overexpression of TagipA, induced by exogenous GT, could indicate a similar regulatory mechanism. These observations corroborate the mounting evidence that SM genes can also be individually expressed in response to specific stimuli. Furthermore, the expression of TagliA∗ and TagipA could indicate that, in GT-non-producing fungi, gliA, and not gliT, is the key gene for self-protection.
Exposure of both fungi to exogenous GT resulted also in the production and secretion of the less toxic bisdethiobis(methylthio)gliotoxin (BmGT). In A. fumigatus, GT is thiomethylated by the methyltransferase GtmA that acts as a post-biosynthetic negative regulator of GT biosynthesis (Dolan et al., 2014). Moreover, GtmA is known to confer resistance to GT via BmGT formation in systems where the gliT-mediated self-protection does not occur as in yeasts (Smith et al., 2016) and A. niger (Manzanares-Miralles et al., 2016). In TaT6776 exposed to the toxin, gtmA was functional. Thus, in Trichoderma GT-non-producing strains, the negative regulatory system represented by gtmA could have instead a role in GT self-protection. Indeed, the maintenance of an active GT self-protection system, based on the action of both gliA and gtmA, could confer them a competitive advantage providing efficient protection against exogenous GT. Actually, as shown by the multiwell growth assay, TaT6776 was able to withstand high concentrations of GT (up to 30 μg/ml), in contrast to other fungi either lacking the gli cluster or without a gliT ortholog (Carberry et al., 2012). As T. gamsii also possesses the same set of related genes but not the GT-BG, it would be interesting to test its ability to self-protect from exogenous GT. The absence or scarce induction of some TvGv29-8 gli genes and of gtmA expression by exogenous GT could reflect the high level of tolerance of this fungus to GT as shown by the growth assay or too permissive conditions.
In conclusion, the Trichoderma GT biological system includes two related genes, the transcription factor gipA and the indirect negative regulator gtmA, in addition to the gli biosynthetic genes (either clustered or sparse) similar to A. fumigatus. Moreover, the GT-BG are very common and polymorphic. On the basis of this circumscribed Trichoderma spp. screening, the most recurrent type is tentatively associated to a GT-non-producing phenotype. TaT6776, ascertained as a GT-non-producing strain with silent GT-BG, activates a gliA∗- and gtmA-mediated self-protection system leading to the synthesis of the less toxic metabolite BmGT.
These findings provide new insights for a safer selection of future BCAs among Trichoderma species, allowing to easily discard those few strains that are able to synthesize GT.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
DB performed real-time-PCR in the presence of exogenous GT, elaborated the real-time PCR data, and drafted the manuscript. LF was involved in the designing of the whole study, and designed and validated the real-time PCR assay and multiwell growth assay. AG set up the HPLC analyses and elaborated the quantification of GT and BmGT in the analyzed samples. MB performed the HPLC analyses. EG designed the study, coordinated the work, performed the genomic analyses of the GT-BG and related genes, and contributed to draft the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Prof. Giovanni Vannacci, University of Pisa, Italy, for kindly providing the strain Trichoderma afroharzianum T6776 analyzed in this study and Dr. Massimo Turina, Ipsp-C.N.R., Italy and Prof. Giuseppe Borsani, University of Brescia, for their helpful suggestions and critical discussion. We also would like to thank Editage (www.editage.com) for the English language editing.
Footnotes
Funding. LF was funded by the “Fondazione Cariplo” and “Regione Lombardia” (Grant Emblematici Maggiori 2015-1080 “La salute della persona: lo sviluppo e la valorizzazione della conoscenza per la prevenzione, la diagnosi precoce e le terapie personalizzate”).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00200/full#supplementary-material
References
- Bayram Ö, Krappmann S., Ni M., Bok J. W., Helmstaedt K., Valerius O., et al. (2008). VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320 1504–1506. 10.1126/science.1155888 [DOI] [PubMed] [Google Scholar]
- Bergmann S., Schümann J., Scherlach K., Lange C., Brakhage A. A., Hertweck C. (2007). Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 3 213–217. 10.1038/nchembio869 [DOI] [PubMed] [Google Scholar]
- Bulgari D., Montagna M., Gobbi E., Faoro F. (2019). Green technology: bacteria-based approach could lead to unsuspected microbe–plant–animal interactions. Microorganisms 7:44. 10.3390/microorganisms7020044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carberry S., Molloy E., Hammel S., O’Keeffe G., Jones G. W., Kavanagh K., et al. (2012). Gliotoxin effects on fungal growth: mechanisms and exploitation. Fungal Genet. Biol. 49 302–312. 10.1016/j.fgb.2012.02.003 [DOI] [PubMed] [Google Scholar]
- Cramer R. A., Gamcsik M. P., Brooking R. M., Najvar L. K., Kirkpatrick W. R., Patterson T. F., et al. (2006). Disruption of a nonribosomal peptide synthetase in Aspergillus fumigatus eliminates gliotoxin production. Eukaryot. Cell 5 972–980. 10.1128/EC.00049-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan S. K., Bock T., Hering V., Owens R. A., Jones G. W., Blankenfeldt W., et al. (2017). Structural, mechanistic and functional insight into gliotoxin bis-thiomethylation in Aspergillus fumigatus. Open Biol. 7:160292. 10.1098/rsob.160292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan S. K., O’Keeffe G., Jones G. W., Doyle S. (2015). Resistance is not futile: gliotoxin biosynthesis, functionality and utility. Trends Microbiol. 23 419–428. 10.1016/j.tim.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Dolan S. K., Owens R. A., O’Keeffe G., Hammel S., Fitzpatrick D. A., Jones G. W., et al. (2014). Regulation of nonribosomal peptide synthesis: bis-thiomethylation attenuates gliotoxin biosynthesis in Aspergillus fumigatus. Chem. Biol. 21 999–1012. 10.1016/j.chembiol.2014.07.006 [DOI] [PubMed] [Google Scholar]
- Druzhinina I. S., Komoń-Zelazowska M., Kredics L., Hatvani L., Antal Z., Belayneh T., et al. (2008). Alternative reproductive strategies of Hypocrea orientalis and genetically close but clonal Trichoderma longibrachiatum, both capable of causing invasive mycoses of humans. Microbiology 154 3447–3459. 10.1099/mic.0.2008/021196-0 [DOI] [PubMed] [Google Scholar]
- Fiorini L., Guglielminetti L., Mariotti L., Curadi M., Picciarelli P., Scartazza A., et al. (2016). Trichoderma harzianum T6776 modulates a complex metabolic network to stimulate tomato cv. Micro-Tom growth. Plant Soil 400 351–366. 10.1007/s11104-015-2736-2736 [DOI] [Google Scholar]
- Gardiner D. M., Howlett B. J. (2005). Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol. Lett. 248 241–248. 10.1016/j.femsle.2005.05.046 [DOI] [PubMed] [Google Scholar]
- Howell C. R., Stipanovic R. D., Lumsden R. D. (1993). Antibiotic production by strains of gliocladium virens and its relation to the biocontrol of cotton seedling diseases. Biocontrol Sci. Technol. 3 435–441. 10.1080/09583159309355298 [DOI] [Google Scholar]
- Keller N. P. (2019). Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol. 17 167–180. 10.1038/s41579-018-0121-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinovas C., Mendes T. A. D. O., Vannier-Santos M. A., Lima-Santos J. (2017). Modulation of human immune response by fungal biocontrol agents. Front. Microbiol. 8:39. 10.3389/fmicb.2017.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek C. P., Herrera-Estrella A., Seidl-Seiboth V., Martinez D. A., Druzhinina I. S., Thon M., et al. (2011). Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 12:R40. 10.1186/gb-2011-12-4-r40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek C. P., Steindorff A. S., Chenthamara K., Manganiello G., Henrissat B., Zhang J., et al. (2019). Evolution and comparative genomics of the most common Trichoderma species. BMC Genomics 20:485. 10.1186/s12864-019-5680-5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanares-Miralles L., Sarikaya-Bayram Ö, Smith E. B., Dolan S. K., Bayram O., Jones G. W., et al. (2016). Quantitative proteomics reveals the mechanism and consequence of gliotoxin-mediated dysregulation of the methionine cycle in Aspergillus niger. J. Proteom. 131 149–162. 10.1016/j.jprot.2015.10.024 [DOI] [PubMed] [Google Scholar]
- Mukherjee P. K., Horwitz B. A., Kenerley C. M. (2012). Secondary metabolism in Trichoderma - a genomic perspective. Microbiology 158 35–45. 10.1099/mic.0.053629-53620 [DOI] [PubMed] [Google Scholar]
- Nützmann H.-W., Scazzocchio C., Osbourn A. (2018). Metabolic gene clusters in eukaryotes. Annu. Rev. Genet. 52 159–183. 10.1146/annurev-genet-120417-131237 [DOI] [PubMed] [Google Scholar]
- Owens R. A., Hammel S., Sheridan K. J., Jones G. W., Doyle S. (2014). A proteomic approach to investigating gene cluster expression and secondary metabolite functionality in Aspergillus fumigatus. PLoS One 9:e106942. 10.1371/journal.pone.0106942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron N. J., Waller R. F., Cozijnsen A. J., Straney D. C., Gardiner D. M., Nierman W. C., et al. (2007). Origin and distribution of epipolythiodioxopiperazine (ETP) gene clusters in filamentous ascomycetes. BMC Evol. Biol. 7:174. 10.1186/1471-2148-7-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor R. H., McCormick S. P., Alexander N. J., Desjardins A. E. (2009). Evidence that a secondary metabolic biosynthetic gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Mol. Microbiol. 74 1128–1142. 10.1111/j.1365-2958.2009.06927.x [DOI] [PubMed] [Google Scholar]
- Reverberi M., Ricelli A., Zjalic S., Fabbri A. A., Fanelli C. (2010). Natural functions of mycotoxins and control of their biosynthesis in fungi. Appl. Microbiol. Biotechnol. 87 899–911. 10.1007/s00253-010-2657-2655 [DOI] [PubMed] [Google Scholar]
- Sandoval-Denis M., Sutton D. A., Cano-Lira J. F., Gené J., Fothergill A. W., Wiederhold N. P., et al. (2014). Phylogeny of the clinically relevant species of the emerging fungus trichoderma and their antifungal susceptibilities. J. Clin. Microbiol. 52 2112–2125. 10.1128/JCM.00429-414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sautour M., Chrétien M. L., Valot S., Lafon I., Basmaciyan L., Legouge C., et al. (2018). First case of proven invasive pulmonary infection due to Trichoderma longibrachiatum in a neutropenic patient with acute leukemia. J. Mycol. Med. 28 659–662. 10.1016/j.mycmed.2018.10.001 [DOI] [PubMed] [Google Scholar]
- Scharf D. H., Brakhage A. A., Mukherjee P. K. (2016). Gliotoxin - bane or boon? Environ. Microbiol. 18 1096–1109. 10.1111/1462-2920.13080 [DOI] [PubMed] [Google Scholar]
- Schoberle T. J., Nguyen-Coleman C. K., Herold J., Yang A., Weirauch M., Hughes T. R., et al. (2014). A Novel C2H2 transcription factor that regulates gliA expression interdependently with GliZ in Aspergillus fumigatus. PLoS Genet. 10:4336. 10.1371/journal.pgen.1004336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Carberry S., Kavanagh K., Haas H., Jones G. W., O’Brien J., et al. (2010). Self-protection against gliotoxin-a component of the gliotoxin biosynthetic cluster, gliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog. 6:952. 10.1371/journal.ppat.1000952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherkhane P. D., Bansal R., Banerjee K., Chatterjee S., Oulkar D., Jain P., et al. (2017). Genomics-driven discovery of the gliovirin biosynthesis gene cluster in the plant beneficial fungus Trichoderma virens. ChemistrySelect 2 3347–3352. 10.1002/slct.201700262 [DOI] [Google Scholar]
- Smith E. B., Dolan S. K., Fitzpatrick D. A., Doyle S., Jones G. W. (2016). Towards understanding the gliotoxin detoxification mechanism: in vivo thiomethylation protects yeast from gliotoxin cytotoxicity. Microb. Cell 3 120–125. 10.15698/mic2016.03.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui J. A., Pardo J., Chang Y. C., Zarember K. A., Nardone G., Galvez E. M., et al. (2007). Gliotoxin is a virulence factor of Aspergillus fumigatus : gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot. Cell 6 1562–1569. 10.1128/ec.00141-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas W. A., Mukherjee P. K., Laughlin D., Wiest A., Moran-Diez M. E., Kenerley C. M. (2014). Role of gliotoxin in the symbiotic and pathogenic interactions of Trichoderma virens. Microbiology 160 2319–2330. 10.1099/mic.0.079210-79210 [DOI] [PubMed] [Google Scholar]
- Wang D. N., Toyotome T., Muraosa Y., Watanabe A., Wuren T., Bunsupa S., et al. (2014). GliA in Aspergillus fumigatus is required for its tolerance to gliotoxin and affects the amount of extracellular and intracellular gliotoxin. Med. Mycol. 52 504–516. 10.1093/mmy/myu007 [DOI] [PubMed] [Google Scholar]
- Weindling R. (1934). Studies on a lethal principle effective in the parasitic action of Trichoderma lignolum on Rhizoctonia solani and other soil fungi. Phytopath 24 1153–1179. [Google Scholar]
- Wilhite S. E., Straney D. C. (1996). Timing of gliotoxin biosynthesis in the fungal biological control agent Gliocladium virens (Trichoderma virens). Appl. Microbiol. Biotechnol. 45 513–518. 10.1007/BF00578464 [DOI] [Google Scholar]
- Wisecaver J. H., Rokas A. (2015). Fungal metabolic gene clusters-caravans traveling across genomes and environments. Front. Microbiol. 6:161. 10.3389/fmicb.2015.00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.