Fig. 3. MYC transactivates WWP1 gene expression toward PTEN suppression.
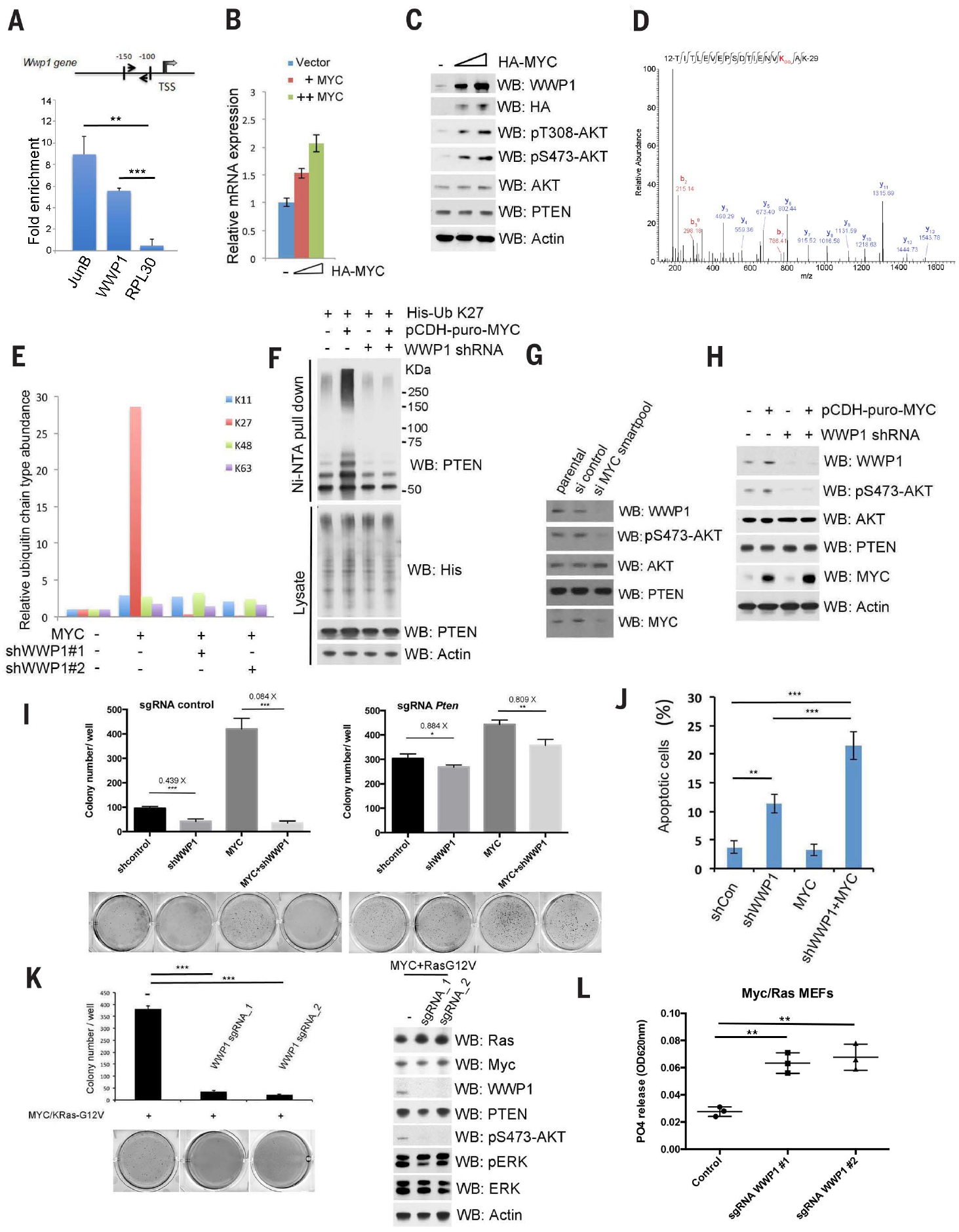
(A) Schematic description of the MYC responsive element on the WWP1 promoter (top). Chromatin level of MYC at the promoter of human WWP1 was measured in DU145 cells. Fold enrichment of MYC was determined by quantitative chromatin immunoprecipitation (qChIP) assays. JunB and RPL30 served as positive and negative controls, respectively. TSS, transcription start site. Data are shown as mean ± SD (***P < 0.0005, **P < 0.005, triplicate experiments, Student’s t test). (B) Reverse transcription–quantitative polymerase chain reaction (RT-qPCR) analysis of WWP1 in DU145 cells expressing the indicated constructs. The mRNA level of WWP1 was determined by RT-qPCR and is presented as a fold increase, as compared with the vector control (−). The HA-MYC levels are indicated by the triangle, from left to right. (C) Analysis of WWP1 and PTEN expression and AKT activation in DU145 cells expressing different amounts of HA-MYC. Total lysates were resolved by SDS-PAGE and then probed with indicated antibodies. The “−” indicates the vector control. (D) Tandem mass spectrum of a peptide derived from endogenous ubiquitinated PTEN in DU145 cells stably expressing the indicated constructs showed ubiquitin conjugation at the K27 residue of ubiquitin. (E) Ratio of indicated ubiquitin linkages detected by MS analysis of endogenous ubiquitinated PTEN purified from MYC overexpression or MYC overexpression–shWWP1 cells to that from control cells (without MYC overexpression). The abundance of each ubiquitin linkage was calculated as described in the materials and methods. (F) Analysis of PTEN K27-linked polyubiquitination in DU145 cells stably expressing MYC and/or WWP1 shRNAs. pCDH-puro-MYC, lentiviral expression of MYC. (G) Analysis of WWP1 and PTEN expression and AKT activation in DU145 cells expressing indicated siRNA SMARTpool. Total lysates were resolved by SDS-PAGE and then probed with indicated antibodies. (H) Analysis of WWP1 and PTEN expression and AKT activation in DU145 cells stably expressing MYC and/or WWP1 shRNAs. (I) Effects of WWP1 with or without PTEN on MYC-induced colony-forming activity in soft agar. The colony numbers are quantified and presented as mean ± SD (***P < 0.0005, **P < 0.005, *P < 0.05, triplicate experiments, Student’s t test). sgRNA, single-guide RNA. (J) Apoptosis assay of DU145 cells stably expressing MYC and/or WWP1 shRNAs. The percentage of apoptotic cells were stained with annexin V–PTEN and 7ADD and then quantified by fluorescence-activated cell sorting. Data are shown as mean ± SD (***P < 0.0005, **P < 0.005, triplicate experiments, Student’s t test). shCon, control. (K) Effects of WWP1 on Ras and MYC-induced colony-forming activity in soft agar or signaling pathway in cells. The colony numbers are quantified and presented as mean ± SD (***P < 0.0005, **P < 0.005, triplicate experiments, Student’s t test) (left). Total lysates were resolved by SDS-PAGE and then probed with indicated antibodies (right). ERK, extracellular signal–regulated kinase; pERK, phosphorylated ERK. (L) Effects of WWP1 on PTEN lipid phosphatase activities in MEFs with indicated constructs. Data are shown as mean ± SD (**P < 0.005, triplicate experiments, Student’s t test). In (C), (F), (H), and (K), actin was used as a loading control.
