FIG 1.
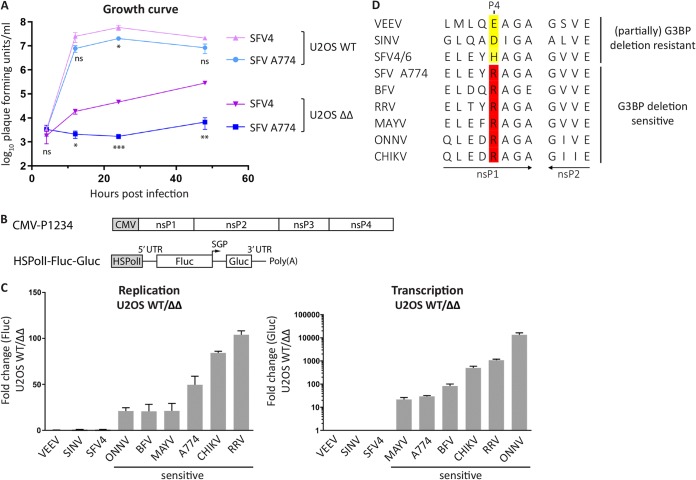
Sensitivity to G3BP deletion correlates with the presence of Arg residue at P4 position of 1/2 site. (A) U2OS WT or ΔΔ cells were infected with SFV4 or SFV A774 at an MOI of 0.1, and at indicated time points, viral titers were quantified by a plaque assay. Data are means of the results from three independent experiments. Error bars indicate the standard error of mean (SEM). SFV A774 virus titers were compared to SFV4 titers at each time point and cell line, and statistical differences were as follows: ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (B) Schematic representation of trans-replication components for expression in human cells. The ns proteins are expressed from a cytomegalovirus (CMV) promoter-based plasmid. The RNA template, which is replicated in trans by the ns proteins, is expressed from a separate plasmid using the human RNA polymerase I promoter (HSPolI). Upon RNA replication, the RNA template resembles the viral RNA but encodes Fluc and Gluc reporters in place of the ns and structural proteins, respectively. SGP, subgenomic promoter; Fluc contains 77 amino acids of nsP1 on its N terminus that are not shown in the figure. UTR, untranslated region. (C) U2OS WT or ΔΔ cells were cotransfected with indicated replicase and corresponding template expression constructs. Fluc (replication) and Gluc (transcription) activities were measured 18 h posttransfection and normalized to signals obtained from control cells transfected with catalytic-inactive replicases and templates. Data are shown as the ratios of replication and transcription signals in U2OS WT to ΔΔ cells from three independent experiments. Means are shown, and error bars indicate the standard deviation (SD). (D) Sequence analysis of 1/2 site residues of replicases analyzed in panel C. Residues at the P4 position are highlighted.