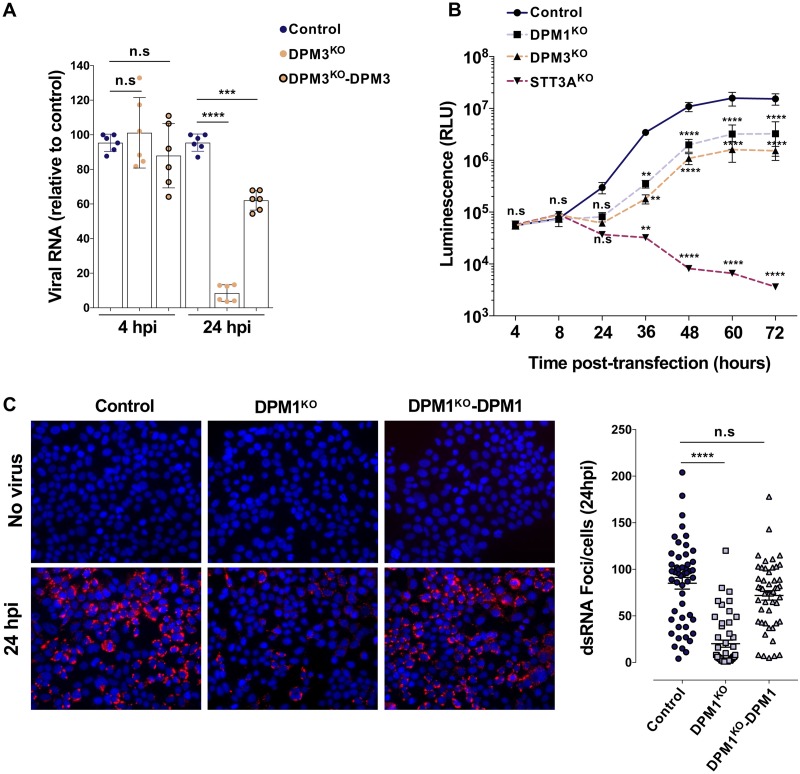
FIG 6.
The DPMS complex is required for optimal viral RNA replication. (A) Control, DPM3KO, and DPM3KO 293T cells complemented with DPM3 were inoculated with DENV 11681 (MOI of 10). At 4 and 24 hpi, cells were treated with trypsin to remove cell surface-bound virus and viral RNA was quantified by RT-qPCR. Data are means ± SD from two independent experiments performed in triplicate. Significance was calculated using a one-way ANOVA with Tukey’s multiple-comparison test. ***, P < 0.001; ****, P < 0.0001). (B) Control, DPM1KO, DPM3KO, and STT3AKO 293T cells were transfected with in vitro-transcribed subgenomic DENV2 RNA expressing the Renilla luciferase. Rluc activity was monitored at the indicated time points. Data are means ± SD from two independent experiments performed in quadruplicate. Significance was calculated using a two-way ANOVA with Dunett’s multiple-comparison test. **, P < 0.01; ****, P < 0.0001. (C) Control, DPM1KO, and DPM1KO 293T cells complemented with DPM1 were inoculated with DENV2 16681 (MOI of 100). Left, representative images of the cells stained with anti-dsRNA MAb J2 at 24 hpi. Right, quantification of the number of foci per cell using Icy software. Data are means ± SD from a representative experiment with n = 50 independent cells. Significance was calculated using a one-way ANOVA with Dunnett’s multiple-comparison test. ****, P < 0.0001.