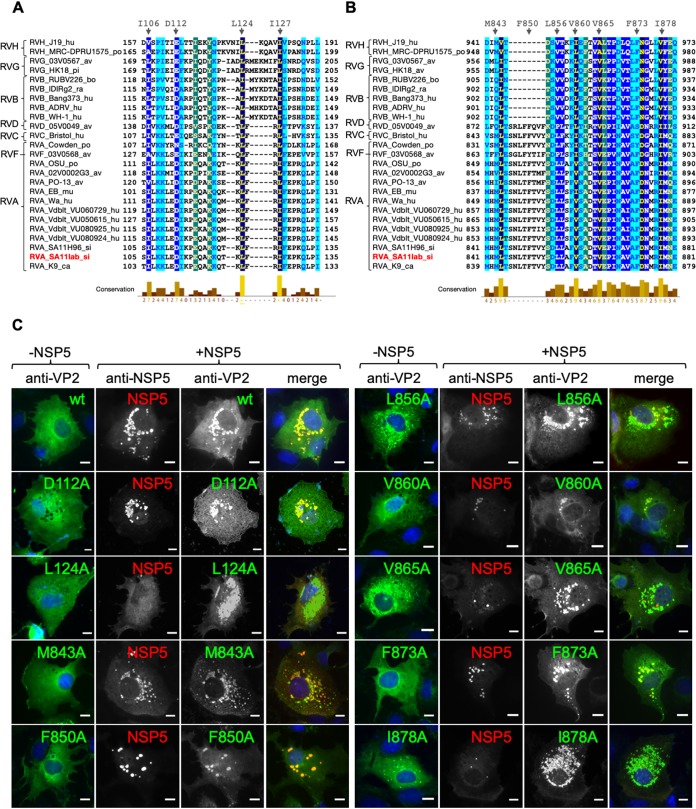
FIG 2.
VP2 residues L124, V865, and I878 are required for VLS formation. Alignment of rotavirus VP2 (strain SA11) amino acid region 103–135 (A) and region 840–880 (B). The VP2 amino acid sequences of several representative virus strains are shown. GenBank accession numbers are listed in Materials and Methods. Members of rotavirus families A to H are indicated. Highly conserved amino acids are shown in dark blue. Amino acid residues mutated to alanine are indicated at the top. A minus sign indicates deletion. (C) Immunofluorescence of MA104 cells expressing wt VP2 or point mutations alone (−NSP5 column) or together with NSP5 (+NSP5 panel). At 16 hpt, cells were fixed and VLS were detected by immunostaining of HA-VP2 (anti-HA, red) or VP2 (anti-VP2, green) with NSP5 (anti-NSP5, green or red). A merged image is shown in the right column. Nuclei were stained with DAPI (blue). Scale bar is 10 μm.