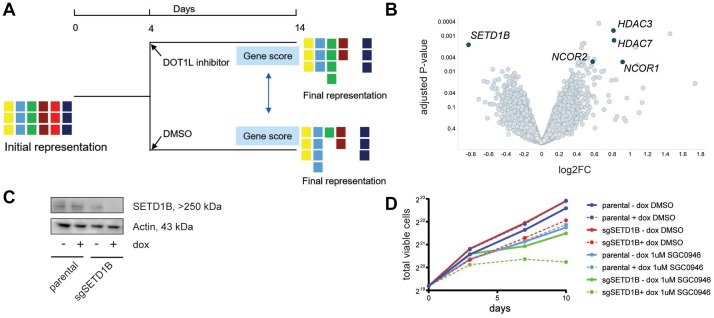
Figure 4. Whole-genome CRISPR screen with or without DOT1L inhibition indicates sensitization by SETD1B inactivation.
(A) Schematic of the screening strategy. MM cells were infected with a pooled lentiviral sgRNA library, selected with puromycin and then divided into two populations: DMSO control and compound treatment (1 μM SGC0946). After 10 days, genomic DNA was isolated from cells, sgRNA sequences were PCR-amplified and abundance of individual sgRNAs was quantified by deep-sequencing. (B) Volcano plot of sensitizers and rescuers in the 4 sensitive cell lines. Each point represents a gene, the x-axis indicates the mean log2-fold changes in sgRNA representation across the 4 cell lines when comparing SGC0946 treatment to DMSO, and the y-axis represents the significance of the log-fold changes (-log10 of P value). (C) Assessment of SETD1B knockout efficiency by western blot in RMPI8226 Cas9 cells upon activation of an inducible sgRNA targeting SETD1B with doxycycline (dox, 100 ng/ml) for 3 days. (D) Effect of SETD1B knockout on growth of RPMI8226 Cas9 cells in the presence or absence of SGC0946. Expression of sgSETD1B was induced with dox as before. Parental RPMI8226 Cas9 cells without sgRNA are shown as control.