Abstract
N6-methyladenosine (m6A), the most abundant and reversible RNA modification, plays critical a role in tumorigenesis. However, whether m6A can regulate p53, a leading antitumor protein remains poorly understood. In this study, we explored the regulatory role of m6A on p53 activation using an arsenite-transformed keratinocyte model, the HaCaT-T cell line. We created the cell line by exposing human keratinocyte HaCaT cells to 1 μM arsenite for 5 months. We found that the cells exhibited an increased m6A level along with an aberrant expression of the methyltransferases, demethylase, and readers of m6A. Moreover, the cells exhibited decreased p53 activity and reduced p53 phosphorylation, acetylation, and transactivation with a high nucleus export rate of p53. Knockdown of the m6A methyltransferase, METTL3 significantly decreased m6A level, restoring p53 activation and inhibiting cellular transformation phenotypes in the arsenite-transformed cells. Further, using both a bioinformatics analysis and experimental approaches, we demonstrated that m6A downregulated the expression of the positive p53 regulator, PRDM2, through the YTHDF2-promoted decay of PRDM2 mRNAs. We showed that m6A upregulated the expression of the negative p53 regulator, YY1 and MDM2 through YTHDF1-stimulated translation of YY1 and MDM2 mRNA. Taken together, our study revealed the novel role of m6A in mediating arsenite-induced human keratinocyte transformation by suppressing p53 activation. This study further sheds light on the mechanisms of arsenic carcinogenesis via RNA epigenetics.
Keywords: N6-methyladenosine, arsenite, p53 activation, cell transformation
Summarizes
Increased m6A attenuates p53 activation in arsenite-transformed human keratinocytes.
Introduction
Arsenite is a widespread environmental pollutant that poses a severe health threat to more than 100 million people worldwide 1,2. It belongs to one of the Class 1 carcinogens designated by the International Agency for Research on Cancer (IARC) 3. However, the carcinogenic mechanisms of arsenite remain to be elucidated. A recent study from our group has demonstrated that N6-methyladenosine (m6A), one of the most abundant and reversible post-transcriptional modifications in eukaryotic RNA, mediates the proliferation and apoptosis of arsenite-transformed human bronchial epithelial cells 4. This suggests a new mechanistic pathway that leads to arsenite carcinogenesis through m6A. This is further supported by the results from the studies showing that m6A is associated with arsenite-induced oxidative stress 5–7, which is the major cause of its carcinogenicity. Increased m6A promoted the proliferation of human keratinocytes that are exposed to low levels of arsenite, possibly through the inhibition of oxidative stress 5. Also, it has been found that arsenite-induced oxidative stress can elevate the level of m6A in keratinocytes 6. All these suggest a new role of m6A in arsenite carcinogenesis. Thus, it is critically important to elucidate the mechanisms by which m6A mediates arsenite-induced carcinogenesis as this can significantly advance our understanding of arsenite carcinogenicity and open a new avenue for prevention of arsenite carcinogenesis.
It has been shown that m6A is also involved in the mutations of the tumor suppressor p53 and regulation of the expression of the genes in the p53 pathway in human cells 8. In myeloid leukemia patients, the genetic alterations of m6A regulatory genes are associated with p53 gene mutation 9. In mice, elevated m6A level alters the expressions of 127 genes, among which 78 genes are in the p53 functional interaction network 10. These suggest a strong association between m6A and activation of the p53 pathway in mammals. Since downregulation of the expression of p53 and its inactivation can mediate arsenite toxicity 11–13, we further hypothesize that m6A mediates arsenite-induced transformation by regulating the expression of p53 and altering its functional activation. To test this hypothesis, in this study, we explored how arsenite can induce cell transformation through the effects of m6A on the p53 expression, activity, and function in human cells using both experimental approaches and bioinformatics analysis of the datasets from several publications.
Given that skin cancer caused by hyperkeratosis is the most common form of cancers induced by arsenite, and cell transformation is a commonly used approach for studying the carcinogenicity of arsenite, we used human keratinocytes that were transformed by chronic arsenite exposure as the model system for this study. We found that m6A mediated arsenite-induced cell transformation by suppressing p53 activation rather than regulating p53 gene expression. In a combination of the results from both the experiments and bioinformatics analysis, we demonstrated that a high m6A level inactivated p53 by modulating the expressions of p53 regulators. Our study provides the first evidence that m6A modification plays a pivotal role in arsenite-induced inactivation of p53, shedding light on a new mechanism by which RNA epigenetics mediates arsenite carcinogenicity.
Materials and Methods
Human keratinocyte culture
The human keratinocyte line (HaCaT) was purchased from KeyGen (Nanjing, China) and cultured by High-glucose Dulbecco’s Modified Eagle’s Medium (DMEM; Life Technologies, USA) with serum in 37 °C humidified incubator containing 5% CO2.
Arsenite treatment and short interfering RNA (siRNAs) transfection
HaCaT cells were continuously maintained in the medium containing 0 or 1 μM sodium arsenite (NaAsO2, Sigma, St. Louis, MO, purity: 99.0%) for 50 passages (about 5 months) with 72 h per passage. HaCaT cells treated by 1 μM arsenite for 50 passages were named as HaCaT-T cells. The transformation phenotypes of HaCaT-T cells were characterized by cell proliferation and clone assays. All short siRNAs were synthesized by RIBOBIO (Guangzhou, China). siR-Ribo™ Negative Control (RIBOBIO, Guangzhou, China) was used as siRNA control (siControl). HaCaT cells and HaCaT-T cells were seeded in 6-well plates with antibiotic-free medium at 2×105 cells/well. Adherent cells were then transfected with METTL3 siRNAs (siMETTL3–1 and siMETTL3–2), YTHDF1 siRNAs (siYTHDF1–1 and siYTHDF1–2), YTHDF2 siRNAs (siYTHDF2–1 and siYTHDF2–2), and negative control (siControl), respectively using riboFECT™ CP (RIBOBIO, Guangzhou, China). The transfected cells were cultured for 72 h and harvested for experiments.
Cell proliferation and viability assay
Cell proliferation and viability were tested by MTT assay. For the determination of cell proliferation rate, HaCaT cells and HaCaT-T cells were cultured in 96-well plates at a density of 2×103 cells/well for 0 h, 24 h, 48 h, 72 h, and 96 h. For the assay on cell viability, 1×104 cells were seeded per well in 96-well plates. Adherent cells were exposed to different concentrations of arsenite (0 μM, 5 μM, 10 μM, 15 μM, 20 μM, 40 μM, 60 μM) for 24 h. Cells were then incubated with MTT (0.5 mg/mL) at 37 °C for 4 h in the dark, and formazan crystals were dissolved by dimethylsulfoxide. Absorbance at 570 nm was measured by Multiskan™GO (Thermo Fisher Scientific, Inc., USA). The cell proliferation rate and viability of each group were normalized to the control, respectively.
Cell anchorage-independent growth measured by soft agar clone formation assay
DMEM with 0.6% agar was used as under-layer soft agar in 96-well plate, 1000 cells in DMEM with 0.35% agar were seeded per well in this plate. After incubated at 37 °C for 2 weeks, cell clones (> 75 μm in diameter) were photographed and counted in a microscope (OLYMPUS, Japan). The clone formation percentage was calculated as follows:
Formation of cell clones detected by plate colony formation assay
One thousand cells were seeded per well in 12-well plates and cultured for 10 days. Cells were then washed twice with phosphate-buffered saline (PBS), fixed in methanol for 20 min, and stained with 10% (w/v) Giemsa for 15 min. Colonies that contain 50 or more cells were counted under a microscope (OLYMPUS, Japan). The colony formation percentage was calculated as follows:
Cell migration determined by wound-healing assay
Cells (1×106) were seeded per well into 6-well plates to form a confluent monolayer. A wound made by a 200 μL pipet tip, and cells were imaged with microscopy after the wound was made 48 h later. The cell migration areas were analyzed by the ImageJ software (National Institutes of Health, USA). The migration area was calculated as follows:
“A0” is the area of the wound at the beginning. “A48” is the area of the wound after 48 h.
Measurement of cell apoptosis by flow cytometry
Apoptosis was detected by Annexin V- fluorescein isothiocyanate (FITC) Apoptosis Detection Kit (Vazyme Biotech Co. Nanjing, China). Floating and attached cells were resuspended in 500 μL of binding buffer, stained with 5 μL propidium iodide (PI) and 5 μL Annexin V-FITC. Cells were protected from light and incubated at room temperature for 30 min. Apoptotic cells (10,000 cells per sample) were immediately analyzed using flow cytometry (BD FACSVerse, US).
Measurement of mRNA levels by quantitative reverse transcription-PCR (RT-qPCR)
Isolation of total RNA, measurement of the amount of RNA, synthesis of cDNA, and detection of RNA levels were performed as described previously 6. The sequences of the PCR primers are listed in Supplementary Table 1. RT-qPCR was performed according to the method described previously 6.
Measurement of protein levels in cells by Western blot
Cells were lysed by lysis buffer (Beyotime, China) containing a phosphatase inhibitor (Servicebio, China) and protease inhibitor, PMSF (Roche, China). Proteins were detected and analyzed according to the method described previously 6. Antibodies against METTL3, METTL14, WTAP, KIAA1429, FTO, YTDHF1, p53 acetyl-lys382, and PRDM2 were from Abcam (Cambridge, UK). Antibodies against p53 and GAPDH were from Proteintech Group (Wuhan, China). Antibodies against p53 phospho-ser15, phospho-ser46, phospho-ser392, MDM2, and YY1 were from Absin Bioscience Inc. (Shanghai, China).
Measurement of total m6A level
The m6A level in total RNA was detected by EpiQuik m6A RNA Methylation Quantification Kit (Epigentek, Farmingdale, USA) as described previously 6. Briefly, RNA samples were incubated with an m6A capture antibody with 1:1000 dilution at room temperature for 1 h, and detection antibody with 1:2000 dilution at room temperature for 30 min. Subsequently, the samples were incubated with the enhancer solution and detection solution at room temperature and subject to the measurement of the absorbance at 450 nm using the Multiskan™ GO plate reader. The amount of m6A was calculated according to the equation described previously 6.
Measurement of p53 activity
The p53 activity in the nuclear extract was detected by the Human p53 Activity Assay Kit (GeneCopoeia, Inc, USA). Nuclear proteins of cells were extracted using Nuclear and Cytoplasmic Protein Extraction Kit (MeiLunbio, China). The concentrations of nuclear proteins were measured by BCA Protein Assay Kit (MeiLunbio, China). Subsequently, p53 activity in nuclear extract was detected by Human p53 Activity Assay Kit (GeneCopoeia, Inc, USA). Briefly, 10 μL sample (50 μg of nuclear proteins) was added into each well of a 96-well plate with 50 μL of Binding Buffer A at room temperature for 1 h with gentle rocking. 100 μL diluted p53 primary antibody was added to each well, and the plate was incubated at room temperature for 1 h. Subsequently, 100 μL diluted HRP conjugated antibody was added to each well, and the plate was incubated at room temperature for an additional 1 h. Then 100 μL TMB Substrate Solution was added to each well, and the plate was incubated at room temperature for 15 min. Finally, 100 μL Stop Solution was added, and the plate was read within 5 min using a Multiskan™ GO plate reader at 450nm.
Localization of p53 protein in cells detected by immunofluorescence
The cellular distribution of p53 was determined by immunofluorescence. Cells (5×105/per well) were seeded in a 6-well plate with a sterilized coverslip at the bottom of each well. Cells were fixed with 4% paraformaldehyde for 15 min and subject to permeabilization using 0.5% Triton X-100 for 20 min. Subsequently, cells were incubated with goat serum for 1 h at 37 °C and a rabbit anti-p53 polyclonal antibody (Proteintech Group, Wuhan, China) at 4 °C overnight. Cells were washed and incubated with a Cy3-conjugated Goat Anti-rabbit IgG (H+L) for 1 h at room temperature. Cells were washed for three times in the dark and incubated with 4′,6-diamidino-2-phenylindole (DAPI) at room temperature for 10 min. Finally, the coverslip was sealed to a slide with an anti-fluorescence quenching agent, and cells were subject to imaging under a fluorescent microscope.
Detection of p53 mutation by DNA sequencing
Genomic DNA of HaCaT and HaCaT-T cells was extracted using Animal Genomic DNA Kit (TsingKe Biotech, China). Exons 5–9 of the p53 gene were amplified by PCR. The sequences of the primers are listed in Supplementary Table 2. PCR was performed under the conditions: 94 °C for 3 min, 1 cycle, denaturation at 94 °C for 30 s, annealing at 58 °C for 3 s, extension at 72 °C for 1 min, 35 cycles. The PCR products of p53 gene in HaCaT and HaCaT-T cells were separated by gel electrophoresis in a 3% low melting agarose gel with EtBr. Subsequently, the PCR products were extracted from the gel, which was melted at 50 °C. The PCR products were reamplified under the same PCR conditions. The PCR products were then cloned into pTZ57R/T vector using InsTAclone PCR Cloning Kit (Thermo Scientific, Canada) and subject to DNA sequencing at the TsingKe Biotech Inc (Beijing, China).
Measurement of the level of m6A in a specific mRNA by m6A RNA immunoprecipitation assay (MeRIP)
The m6A levels in the mRNA of the p53, PRDM2, MDM2, and YY1 gene were determined using Magna MeRIP™ m6A Kit (A&D Technology Corporation, Beijing, China). Total RNA was extracted and analyzed as described above. mRNA was purified from total RNA using the Oligotex mRNA Kits (QIAGEN Group Inc., USA), and fragmented into 100 bp or smaller fragments. These fragmented mRNAs were further purified using miRcute miRNA isolation kit (Tiangen Biotech, Beijing Co., Ltd, China). Fragmented mRNAs were transferred into a new microcentrifuge tube followed by magnetic immunoprecipitation with m6A monoclonal antibody. Briefly, MeRIP reaction mixture was incubated with an anti-m6A antibody attached to magnetic beads for 2 h at 4 °C. The level of m6A in the antibody-precipitated RNA fragments was determined by RT-qPCR. The levels of m6A in the indicated genes were normalized to that of the internal control, GAPDH.
Bioinformatics analysis
Differentially expressed genes between HaCaT cells and HaCaT cells through chronic exposure to low concentrations of arsenite were obtained by GSE97303 13. The correlation among these differentially expressed genes, p53, and m6A-regulating proteins was analyzed by STRIGN (https://string-db.org/). The sites of m6A in mRNA were determined by analyzing m6AVAR database (http://m6avar.renlab.org/index.html) 14 An analysis of the gene expression in skin cutaneous melanoma (SKCM) and its tumor purity, i.e., the percentage of cancer cells in total cells of tumor tissue, was performed using LINKEDOMICS (http://www.linkedomics.org/admin.php). The correlation of the gene expression with SKCM stage and prognosis was analyzed using GEPIA (http://gepia.cancer-pku.cn/index.html). The data of the gene expression in the cells and animals that were exposed to arsenic were obtained from GEO database (GSE47047, GSE97303, GSE103873, GSE2187, GSE33520, GSE36684, and GSE21193). Mutation and gene network analysis was performed using CBIOPORTAL (https://www.cbioportal.org/) and GENEMANIA (https://genemania.org/). The m6A peaks on the PRDM2, MDM2, and YY1 mRNA were analyzed using CVm6A (http://gb.whu.edu.cn:8080/CVm6A).
Statistical analysis
All results were illustrated as mean ± standard error of the mean (SEM). A statistical significance was detected using Student’s t-test or one-way analysis of variance (ANOVA) with SPSS 19.0 (SPSS Inc., Chicago, IL, USA). Bar charts and heat maps were created using the GraphPad Prism 7.0 software. The interactive reticular chart was obtained by Cytoscape 15. P < 0.05 was designated as a statistical difference.
Data availability
The source data for Figs. 1–6 are provided in Supplementary Table 1–5. The source data for Supplementary Figs. 1–6 are provided in Supplementary Table 6. The source data for p53 sequence are provided in the Supplementary p53 sequence.
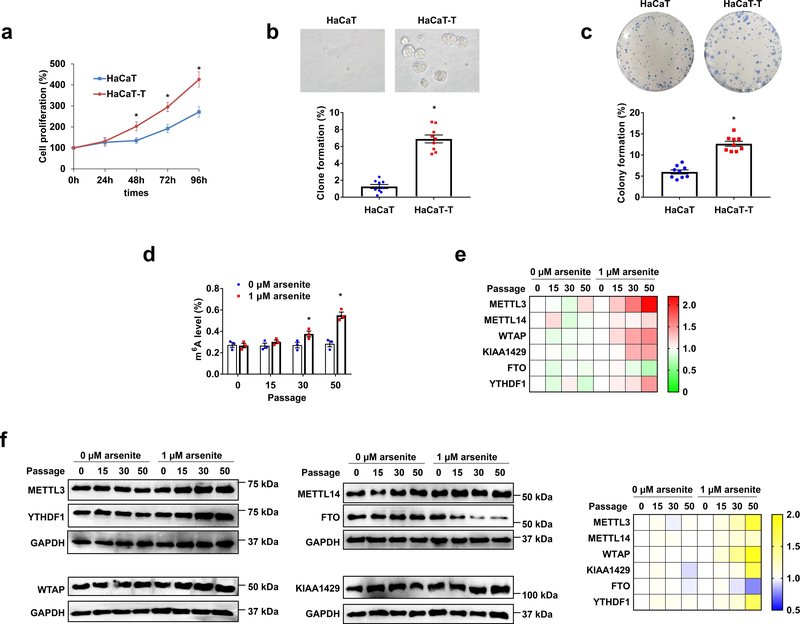
Fig. 1. m6A level increases in arsenite-transformed human keratinocytes.
a, Cell proliferation of HaCaT and HaCaT-T cells was measured by MTT assay. b, Percentage of clone formation was calculated by soft agar clone formation assay in HaCaT and HaCaT-T cells (down). Representative images of HaCaT and HaCaT-T cells in soft agar clone formation assay observed with a microscope at 400× magnification (up). c, Percentage of colony formation was tested by plate colony formation assay in HaCaT and HaCaT-T cells (down). Representative images of HaCaT and HaCaT-T cells in plate colony formation (up). d, Total m6A levels in HaCaT and HaCaT-T cells of 0, 15, 30, 50 passages were determined by EpiQuik m6A RNA Methylation Quantification Kit. e, f, RT-qPCR testing the mRNA levels (e) and western blotting examining the protein levels (f) of m6A regulating proteins (METTL3, METTL14, WTAP, KIAA1429, FTO, and YTHDF1) in HaCaT and HaCaT-T cells of 0, 15, 30, 50 passages are shown in heat maps. Representative images of immunoblot (f, left). For a-f, n = at least 3 biological replicates, error bars indicate mean ± SEM. The P-values were determined by two-tailed t-test. “*” indicates a significant difference compared to control group (P < 0.05).
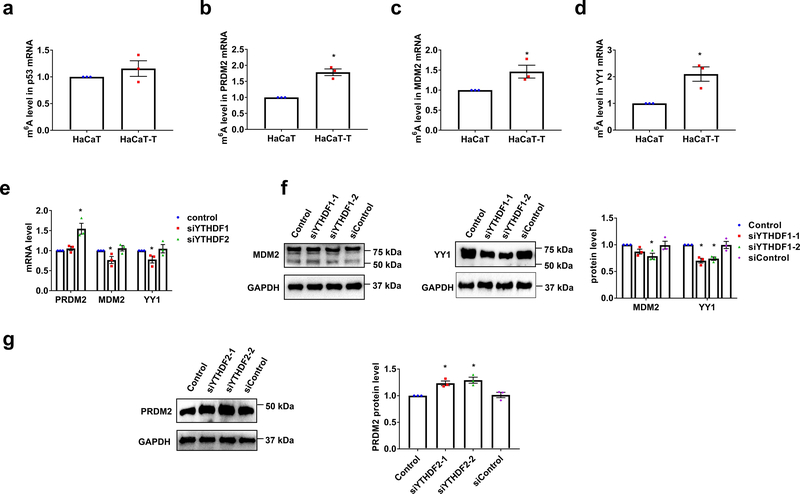
Fig. 6. m6A methylation inhibits PRDM2 expression and increases MDM2 and YY1 levels in HaCaT-T cells.
a-d, m6A levels in p53 (a), PRDM2 (b), MDM2 (c), and YY1 (d) mRNA of HaCaT and HaCaT-T cells were determined by MeRIP. e, The mRNA levels of PRDM2, MDM2, and YY1 in HaCaT-T cells with introduction of YTHDF1 siRNA (siYTHDF1) or YTHDF2 siRNA (siYTHDF2) were tested by RT-qPCR. f, The representative images of MDM2 (left) and YY1 (middle) in HaCaT-T cells with introduction of two YTHDF1 siRNA (siYTHDF1–1 and siYTHDF1–2). Quantitative protein levels of MDM2 and YY1 in HaCaT-T cells with introduction of two YTHDF1 siRNA were detected by western blotting (right). g, The representative images (left) of PRDM2 in HaCaT-T cells with introduction of two YTHDF2 siRNA (siYTHDF2–1 and siYTHDF2–2). Quantitative protein levels of PRDM2 in HaCaT-T cells with introduction of two YTHDF2 siRNA were detected by western blotting (right). For a-g, n = 3 biological replicates, error bars indicate mean ± SEM. The P-values were determined by two-tailed t-test. “*” indicates a significant difference compared to control group (P < 0.05).
Results
The level of m6A is increased in arsenite-transformed human keratinocytes, HaCaT-T cells
To determine if the level of m6A may be altered in arsenite-transformed human keratinocytes, we initially established an arsenite-transformed human keratinocyte line by chronically exposing human HaCaT keratinocytes to 1 μM arsenite for 5 months (50 passages). The transformed cell line was designated as HaCaT-T cells. We then characterized the transformation phenotypes of HaCaT-T cells. Compared with HaCaT cells (passed for 50 passages without arsenic exposure), HaCaT-T cells exhibited increased cell proliferation with a higher cell proliferation percentage than HaCaT cells at the time points of 48 h - 96 h (Fig. 1a). Also, HaCaT-T cells exhibited a significantly increased capacity of forming cell clones compared with HaCaT cells in both soft agar and 12-well plates (Fig. 1b and 1c). The results indicate that HaCaT-T cells were successfully transformed by chronic arsenite exposure.
To further determine the changes in the level of m6A during arsenite-induced transformation, we measured the level of m6A in HaCaT cells that were chronically exposed to a low dose of arsenite (1 μM). We found that the level of m6A was significantly increased in arsenite-treated HaCaT cells at 30 and 50 passages (Fig. 1d). Since the level and function of m6A are modulated by m6A methyltransferases, demethylases, and m6A readers 16–18, we further examined the expression levels of the m6A-regulatory proteins in the arsenite-treated HaCaT cells at different passage numbers (Fig. 1e and 1f). We found that the expression of the m6A methyltransferases, METTL3, METTL14, WTAP, KIAA1429, and the m6A reader, YTHDF1 was increased in arsenite-treated HaCaT cells compared with the untreated HaCaT cells (Fig. 1e). Along with the increasing passage numbers, the expression of the proteins increased (Fig. 1f). In contrast, the demethylase of m6A, FTO was decreased in the arsenite-treated cells at passage 30–50 (Fig. 1e and 1f). Our results are consistent with those from our bioinformatics analysis of the dataset from the TCGA database showing that the mRNA levels of METTL3, METTL14, KIAA1429, YTHDF1, and YTHDF2 are all positively associated with the tumor purity of SKCM (Supplementary Fig. 1a). In addition, a higher level of METTL3 and WTAP in the patients of SKCM of stage I-IV than that in stage 0 was detected (Supplementary Fig. 1b). All these results suggest that an increased level of m6A induced by arsenite is associated with human keratinocyte transformation and skin cancer. Thus, it is possible that m6A level in the cells may be altered by the aberrant expressions of m6A methyltransferases, demethylases, and readers induced by arsenite resulting in keratinocyte transformation.
m6A mediates the `migration and apoptosis of arsenite-transformed keratinocytes, HaCaT-T cells
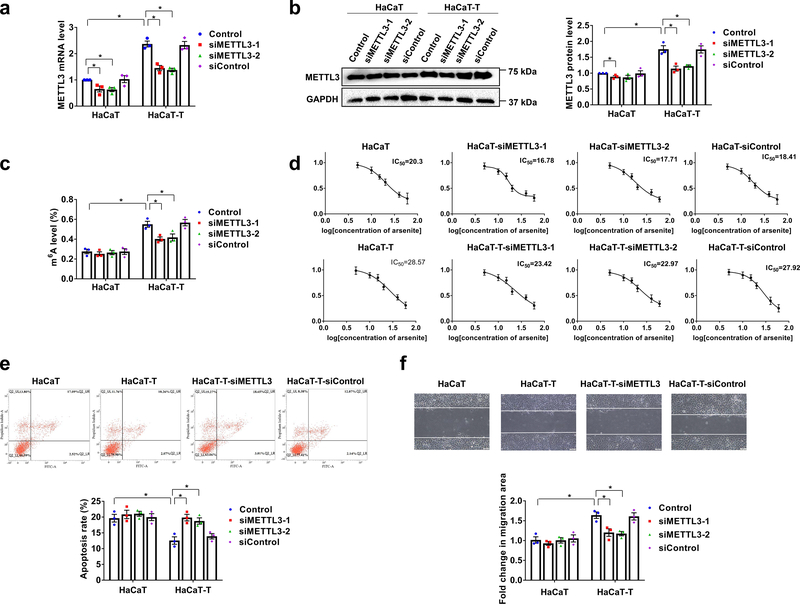
To further determine the effects of m6A methylation on arsenite-induced transformation, we examined the viability, migration, and apoptosis of HaCaT-T cells under the downregulation of m6A by knocking down METTL3 using siRNAs (Fig. 2). We initially validated knockdown of METTL3 and the reduction of m6A in HaCaT-T cells. As shown in Fig. 2a and 2b, knockdown of METTL3 significantly reduced its mRNA and protein levels in HaCaT cells and HaCaT-T cells (Fig. 2a and 2b). Consequently, the m6A level in total RNA was significantly decreased compared with the HaCaT-T cells without or with scrambled siRNA (siControl) (Fig. 2c). However, the m6A level in HaCaT cells was not altered by the knockdown of the methyltransferase (Fig. 2c). The results showed that METTL3 knockdown specifically reduced the level of METTL3 and m6A in HaCaT-T cells. We then determined if this further affected the sensitivity of HaCaT-T cells to arsenite (Fig. 2d). We found that arsenite was more effective in reducing the viability of HaCaT-T cells with siMETTL3 than that of HaCaT-T cells without the siRNAs and the siControl (Fig. 2d). Next, we examined if METTL3 knockdown facilitated cell death through apoptosis induced by the cellular exposure to10 μM arsenite for 24 h (Fig. 2e). Indeed, we found that METTL3 knockdown resulted in a higher apoptosis rate in HaCaT-T cells than HaCaT-T cells and the siControl (Fig. 2e). The apoptosis rate was comparable to that of HaCaT with or without siRNAs (Fig. 2e, the bar chart). Finally, we examined the effects of METTL3 knockdown on the migration HaCaT-T cells using the wound-healing assay. The results revealed that HaCaT-T cell migration was significantly inhibited by METTL3 knockdown compared with the control cells (Fig. 2f). The results are consistent with those from our analysis of GEO database demonstrating that chronic arsenic exposure increased the expression of other m6A methyltransferases, METTL3, METTL14, WTAP, KIAA1429, and the m6A reader, YTHDF1 and decreased the expression of m6A demethylase, FTO in several cell lines and animal models (Supplementary Fig. 2). This further indicates a possible role of m6A in arsenite-induced cell transformation.
Fig. 2. m6A mediates migration and apoptosis in arsenite-transformed keratinocytes.
a, b, The mRNA (a) and protein (b) levels of METTL3 in HaCaT and HaCaT-T cells with introduction of two siMETTL3 (siMETTL3–1 and siMETTL3–2) and siControl were tested by RT-qPCR and western blotting, respectively. Representative images of western blotting (b, left). c, Total m6A levels in HaCaT and HaCaT-T cells with introduction of siMETTL3 and siControl were determined by EpiQuik m6A RNA Methylation Quantification Kit. d, Cell viability of HaCaT and HaCaT-T cells with different treatments was tested by MTT assay. e, Representative images of apoptosis tested by flow cytometry (up) and quantification of cell apoptosis in HaCaT cells and HaCaT-T cells with induction of two siMETTL3 (siMETTL3–1 and siMETTL3–2) and siControl (down). f, Representative images of cell migration examined by wound healing assay with a microscope at 400× magnification (up) and quantification of migration in HaCaT cells and HaCaT-T cells with induction of two siMETTL3 (siMETTL3–1 and siMETTL3–2) and siControl (down). For a-f, n = 3 biological replicates, error bars indicate mean ± SEM. The P-values were determined by two-tailed t-test. “*” indicates a significant difference compared to control group (P < 0.05).
m6A attenuates p53 activation to mediate arsenite-induced transformation.
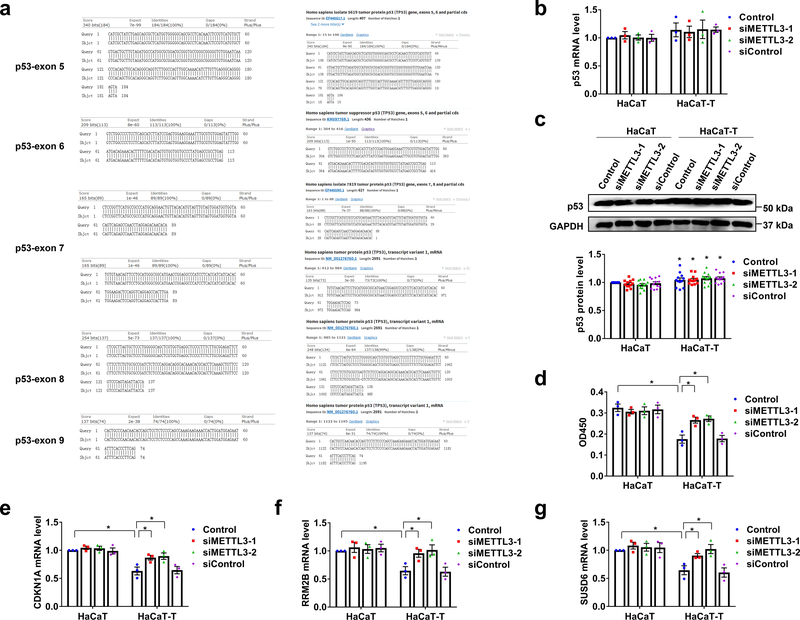
Since m6A is associated with p53 function 8–10, we further hypothesize that m6A mediates arsenite-induced transformation by regulating the expression and activation of p53. To test this possibility, we initially determined if the mutations of p53 can be involved in arsenite-induced keratinocyte transformation, we sequenced a segment in the p53 encoding region of HaCaT and HaCaT-T cells (exons 5–9, commonly mutated regions in cancers 19) (Fig. 3a). We failed to detect any mutations and differences in the sequence of the regions between HaCaT and HaCaT-T cells (Fig. 3a). The results are consistent with those from our bioinformatics analysis of the TCGA database showing that p53 mutations did not affect the survival of SKCM patients (Supplementary Fig. 3a) and are not associated with the expressions of m6A-related proteins in SKCM tissue (Supplementary Fig. 3b). This further suggests that p53 mutations were not involved in m6A-mediated HaCaT transformation induced by arsenite.
Fig. 3. p53 mutation, expression, activity, and target genes level in arsenite-transformed keratinocytes.
a, The sequence of p53 gene (exons 5–9) in HaCaT and HaCaT-T cells was detected by sequencing. b-d, The mRNA (b) and protein (c) levels and activity (d) of p53 in HaCaT and HaCaT-T cells after the indicated treatments. Representative images of western blotting (c, up). e-g, The mRNA levels of three p53 specific target genes including CDKNA1 (e), RRM2B (f), and SUSD6 (g) were examined by RT-qPCR. For b-g, At least 3 biological replicates, error bars indicate mean ± SEM. The P-values were determined by two-tailed t-test. “*” indicates a significant difference compared to control group (P < 0.05).
To further determine if m6A can affect the expression level of p53, we examined the expression of p53 in HaCaT and HaCaT-T cells with METTL3 knockdown (Fig. 3b and 3c). We found that the mRNA level of p53 in these cells was not significantly altered by the METTL3 knockdown. Instead, the gene knockdown did not result in a higher p53 protein level in HaCaT-T cells than HaCaT cells (Fig. 3c), suggesting that m6A mediated arsenite-induced cell transformation independent of p53 gene expression. This is further supported by our analysis of the GEO database showing that the mRNA level of p53 in HaCaT, prostate epithelial cells, and rat liver tissue was not significantly altered by arsenite (Supplementary Fig. 3c).
We then asked if m6A can mediate arsenite-induced keratinocyte transformation by modulating the activation of p53. We found that p53 activity was significantly decreased in HaCaT-T cells, and this was reversed by knockdown of METTL3 in HaCaT-T cells (Fig. 3d). Since p53 activity can also be determined by the expression of the p53 target genes, the cellular localization and posttranslational modifications of p53 protein 20–22, we then examined the expression of three p53 target genes, CDKN1A, RRM2B, SUSD6 (KIAA0247) in HaCaT and HaCaT-T cells. These genes are involved in cell cycle arrest in G1 phase 23, DNA repair and mtDNA synthesis 24, suppression of cell proliferation, and angiogenesis and promotion of apoptosis 25. We found that all the p53 target genes were significantly lower in HaCaT-T cells than HaCaT cells (Fig. 3e–3g). The suppression of the gene expression in HaCaT-T cells was relieved by knockdown of METTL3 (Fig. 3e–3g). In consistency with our experimental results, the results from our bioinformatics analysis showed that the expression of RRM2B, CDKN1A and SUSD6 was also decreased in HaCaT cells and mice (Supplementary Fig. 4a), SKCM cancer tissue (Supplementary Fig. 4b), and patients (Supplementary Fig. 4c and 4d). All these results suggest that m6A mediated arsenite-induced keratinocyte transformation by reducing the transcriptional activity of p53.
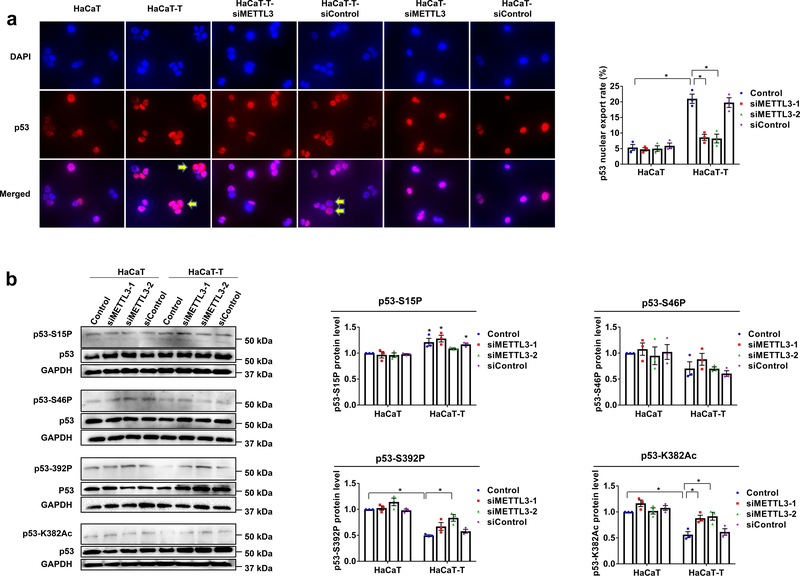
We then determined if m6A can promote the nuclear export of p53 protein to inactivate the protein by altering its cellular compartmentalization 11. We found that in HaCaT-T cells, p53 nuclear localization was significantly reduced along with an increased amount of cytoplasmic staining of p53, and this was significantly reversed by knockdown of METTL3 in HaCaT-T cells (Fig. 4a). To further determine the roles of m6A on p53 activation in mediating arsenite-induced HaCaT cell transformation, we also measured the level of p53 phosphorylation and acetylation at different amino acids (Fig. 4b). We found that the phosphorylation of p53 at Ser15 that is critical for p53 stabilization 26 was increased in HaCaT-T cells (Fig. 4b). Also, we found that p53 phosphorylation at Ser392 that is essential for the stability of p53 tetramerization domain and its DNA binding 27,28, was significantly decreased in HaCaT-T cells (Fig. 4b), and p53 acetylation at Lys382 that is critical for p53 activation 29 was reduced in the cells (Fig. 4b). Further, we found that knockdown of METTL3 significantly increased the phosphorylation at Ser392 and acetylation at Lys382 in HaCaT-T cells (Fig. 4b). However, we found that elevated phosphorylation at Ser15 was not ameliorated in HaCaT-T cells with siMETTL3 (Fig. 4b). The results suggest that m6A mediated arsenite-induced human keratinocyte transformation by suppressing p53 activation rather than affecting p53 gene expression.
Fig. 4. m6A methylation reduces p53 activation in HaCaT-T cells.
a, The localization of p53 protein (red) and nucleus (blue) detected by immunofluorescence was observed with a fluorescent microscope at 400× magnification and merged by Adobe Photoshop. Cytoplasmic p53 staining is depicted by the yellow arrows (left). Quantification of p53 nuclear export rates (right). b, Phosphorylation levels of p53 at Ser15 site, Ser46 site, Ser392 site and p53 acetylation at Lys382 were determined by western blotting in HaCaT and HaCaT-T cells after the indicated treatments. The representative images of protein levels in p53-S15P, p53-S46P, p53-S392P, p53-K382Ac, p53 and GAPDH (left). For a, b, n = 3 biological replicates, error bars indicate mean ± SEM. The P-values were determined by two-tailed t-test. “*” indicates a significant difference compared to control group (P < 0.05).
m6A modulates the expression of p53 activation regulators to mediate arsenite-induced keratinocyte transformation
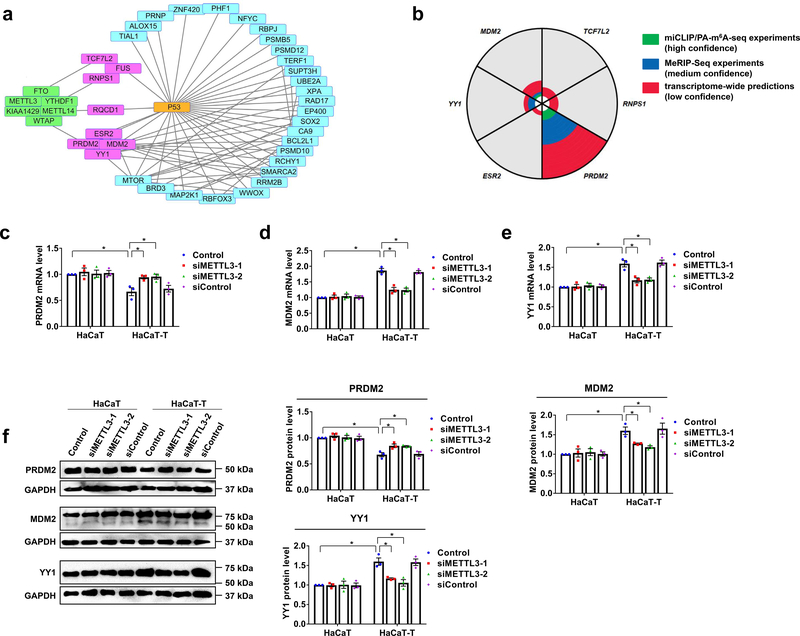
To further explore the mechanisms by which m6A inhibited p53 activation, we initially conducted a bioinformatics analysis of the published data (GSE 97303) to identify the genes that are associated with m6A and p53 in HaCaT cells that were chronically exposed to a low dose of arsenite. We found that the PRDM2, MDM2, YY1, ESR2, TCF7L2, FUS, RNPS1, and RQCD1 gene that are involved in arsenite-induced transformation, were associated with m6A and p53 (Fig. 5a, boxes in pink; Supplementary Table 3). Among these genes, the PRDM2, MDM2, and YY1 gene, which are the regulators of p53 activation 30–32 have the m6A sites in their mRNAs, which have been identified by the miCLIP/PA-m6A-seq or MeRIP-Seq experiments (Fig. 5b; Supplementary Table 4). Our experimental results further demonstrated that the mRNA and protein levels of PRDM2 that upregulates p53 activation 30 were decreased in HaCaT-T cells compared to HaCaT cells (Fig. 5c and 5f). On the other hand, we found that in HaCaT-T cells, the mRNA and protein levels of MDM2, an inhibitor of p53 activation 31,33,34 and YY1, a negative regulator of p53 32 were increased (Fig. 5d–5f). The changes of PRDM2, MDM2, and YY1 are consistent with p53 inactivation in HaCaT-T cells. Moreover, our bioinformatics analysis found that the expression of PRDM2 was also decreased in arsenite-transformed malignant prostate epithelial cells, and the expression of YY1 was increased in human bronchial epithelial cell line and mice that were chronically exposed to arsenite (Supplementary Fig. 5a). Furthermore, we found that the mRNA level of PRDM2 was negatively correlated to SKCM tumor purity, whereas the mRNA level of MDM2 and YY1 was positively correlated to SKCM tumor purity (Supplementary Fig. 5b). Our bioinformatics analysis also showed that the YY1 RNA level was elevated in SKCM patients at stage I-IV (Supplementary Fig. 5c) and served as the core of the network, including MDM2 and p53 (Supplementary Fig. 5d). All of these results indicate that the p53 activation regulators affected by m6A were involved in arsenite-induced transformation of human keratinocytes.
Fig. 5. Three p53 regulators are affected by m6A methylation in HaCaT-T cells.
a, Protein relation analysis on m6A regulating proteins, p53, and genes with altered expressions in HaCaT cells after chronically exposed to low dose of NaAsO2 (GSE97303) was performed by String. The genes in pink and blue rectangles differentially express in HaCaT cells chronically exposed to NaAsO2 compared to HaCaT cells. The proteins in green rectangles are m6A regulating proteins. The genes in pink rectangles are involved in NaAsO2-induced malignant transformation and associated with m6A methylation and p53. Straight lines represent two molecules are close related. b, The analysis of m6A sites in genes which are involved in NaAsO2-induced malignant transformation and associated with m6A and p53 was performed in m6AVAR. The “green” means there are m6A sites in this gene from miCLIP/PA-m6A-seq experiments with high confidence. The “blue” indicates there are m6A sites in this gene from MeRIP-Seq experiments which is medium confidence. The “red” suggests there are m6A sites in this gene from transcriptome-wide predictions with low confidence. The area of “green”, “blue”, or “red” represents the relative counts of m6A sites in this gene. c-e, The mRNA levels of three genes including PRDM2 (c), MDM2 (d), and YY1 (e) were tested by RT-qPCR in HaCaT and HaCaT-T cells with introduction of two siMETTL3 (siMETTL3–1 and siMETTL3–2) and siControl. f, The representative images of PRDM2, MDM2, YY1, and GAPDH protein (left). Quantitative protein levels of PRDM2, MDM2, and YY1 protein in HaCaT and HaCaT-T cells with introduction of METTL3 siRNA were detected by western blotting (right). For c-f, n = 3 biological replicates, error bars indicate mean ± SEM. The P-values were determined by two-tailed t-test. “*” indicates a significant difference compared to control group (P < 0.05).
To further determine if the expression of PRDM2, MDM2, and YY1 can be modulated by elevated m6A, we performed an additional bioinformatic analysis of the dataset from the TCGA database, the results showed that the mRNA level of these p53 regulators was significantly correlated to that of METTL3 gene in SKCM patient tissue (Supplementary Fig. 6a) suggesting that m6A stimulated the expression of the p53 activation regulators in the patient tissue. To determine if m6A is involved in the arsenite-induced keratinocyte transformation through the regulation of the expression of the PRDM2, MDM2, and YY1 gene, we then examined if knockdown of METTL3 can alter the expression of the genes in HaCaT-T cells. We found that knockdown of METTL3 by METTL3 siRNA (siMETTL3) in HaCaT-T cells partially relieved the inhibition of the p53 positive regulator, PRDM2, but decreased the level of the overexpression of the p53 suppressor, MDM2 and YY1 in the cells (Fig 5c–5f). Furthermore, since our bioinformatic analysis of the m6A database (CVm6A) showed m6A peaks on the mRNA of the PRDM2, MDM2, and YY1 (Supplementary Fig. 6b), we then determined the m6A level in p53 mRNA in HaCaT and HaCaT-T cells using MeRIP. The results showed that although the m6A level in p53 mRNA was not significantly altered in HaCaT-T cells (Fig. 6a), the m6A levels on the mRNA of the PRDM2, MDM2, and YY1 were significantly increased in HaCaT-T cells compared with HaCaT cells (Fig. 6b–6d). These results suggested that elevated m6A regulated p53 activation by mediating the regulators of p53 activation in arsenite-induced human keratinocyte transformation.
To explore the mechanisms by which m6A modulates the regulators of p53 activation in HaCaT-T cells, we knocked down YTHDF1 and YTHDF2 in the cells using siRNAs. We found that knockdown of YTHDF1 that promotes translation of m6A-methylated transcripts decreased both the mRNA and protein levels of MDM2 and YY1 (Fig. 6e and 6f). However, knockdown of YTHDF2 that promotes the decay of m6A-methylated transcripts, elevated the mRNA and protein levels of PRDM2 in the cells (Fig. 6e and 6g). The results indicated that decreased YTHDF1 suppressed the expression of MDM2 and YY1, whereas decreased YTHDF2 increased the level of PRDM2 mRNA and protein. Taken together, both our bioinformatics analysis and experimental results revealed that m6A resulted in p53 inactivation by modulating p53 activation regulators in arsenite-transformed human keratinocytes.
Discussion
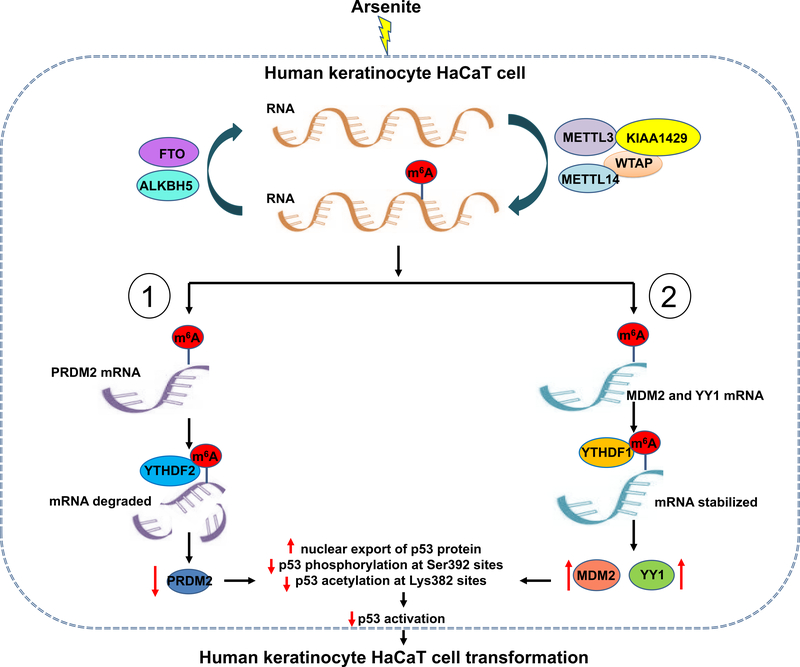
In this study, we discovered that in the arsenite-transformed keratinocytes HaCaT-T cells, m6A methyltransferases, METTL3, METTL14, WTAP, and KIAA1429 were upregulated, whereas the m6A demethylase, FTO was downregulated (Figure 1). We demonstrated that the cellular level of m6A was increased (Figure 1d), and this, in turn, suppressed the expression of the upregulator of p53 activation, PRDM2, and stimulated the expression of the downregulators of p53 activation, MDM2, and YY1 (Figure 5–6). Consequently, this led to p53 inactivation and cellular transformation of human keratinocytes. Our results suggest that m6A mediated arsenite-induced transformation of keratinocytes through suppression of p53 activation. Our results further support a model during which arsenite stimulates the RNA methyltransferases and cofactors, METTL3, METTL14, KIAA1429, and WTAP (Fig. 7). This induces the production of m6A in the mRNAs of PRDM2 (Fig. 7, pathway 1) or that of m6A in the mRNAs of MDM2 and YY1 (Fig. 7, pathway 2). Subsequently, m6A in the mRNAs of PRDM2 is bound by YTHDF2, whereas m6A in the mRNAs of MDM2 and YY1 is bound by YTHDF1 resulting in the degradation of PRDM2 mRNA (Fig. 7, pathway 1) and stabilization of the mRNA of MDM2 and YY1 (Fig. 7, pathway 2). Consequently, this increases p53 nuclear export and decreases p53 phosphorylation and acetylation at Ser392 and Ser382, respectively, leading to p53 inactivation and keratinocyte transformation (Fig. 7).
Fig. 7. Model that elevated m6A methylation inhibits p53 activation to mediate arsenite-induced human keratinocytes transformation.
Chronical exposure of arsenite induces the overexpression of m6A methyltransferases (METTL3, METTL14, WTAP, and KIAA1429) and the attenuation of m6A demethylases (FTO), leading to elevated m6A methylation level in transformation of human keratinocytes. Upregulated m6A methylation in p53 positive regulator PRDM2 mRNA binds with m6A reader YTHDF2, inducing the degradation of PRDM2 mRNA and inhibition of PRDM2 expression. Whereas high levels of m6A methylation in p53 negative regulator MDM2 and YY1 mRNA combines with m6A reader YTHDF1, promoting the stabilization and translation of MDM2 and YY1 mRNA. The suppression of p53 positive regulator and the increase of p53 negative regulators induced by m6A methylation contribute to p53 inactivation involved in arsenite-caused transformation.
Among several proteins that have been identified in the m6A “writer” complex, including METTL3, METTL14, WTAP, and KIAA1429 16–18, METTL3 is the one that plays a key role in generating m6A. This is because genetic depletion of METTL3 in plants, yeast, and mammalian cells results in nearly complete loss of m6A in polyadenylated RNA 34–36. However, other members in the complex do not possess catalytic activity and are only involved in stabilizing the assembly of the complex and recruitment of METTL3 to its target mRNAs in specific subcellular locations 37. Also, METTL3 is overexpressed in different types of cells, including HaCaT cells and animals that are exposed to arsenite based on our experimental results (Fig. 1e and 1f) and our bioinformatics analysis of the published databases (Supplementary Fig. 2). Moreover, our analysis of the TCGA database showed that the mRNA level of METTL3 is positively associated with tumor purity of skin cutaneous melanoma (Supplementary Fig. 1a), and a higher level of METTL3 was detected in the patients of skin cutaneous melanoma of stage I-IV than those in stage 0 (Supplementary Fig. 1b). These results indicate a crucial role of METTL3 in m6A production and regulation and arsenite-induced skin carcinogenesis. Therefore, we decided to knockdown METTL3 in our experiments. Interestingly, we did not observe a significant change in m6A level in HaCaT cells with knockdown of METTL3. This may be due to a low m6A level in HaCaT cells. It has been reported that approximately 0.1–0.4% of adenosine nucleotides in mammalian RNAs are chemically modified 38. In our experiments, m6A level was only about 0.275% in HaCaT cells. Our results showed that knockdown of METTL3 decreased the m6A level in HaCaT cells from 0.275% to 0.255%. Such a small change of the m6A level was not statistically significant.
Previous studies have focused on the mechanisms underlying arsenite carcinogenesis through aberrant gene expression and mutations, as well as altered functions of proteins. Here, we proposed a new mechanism by which arsenite induces cell transformation associated with skin cancer through m6A. A recent study from our group has provided the first evidence that m6A is involved in arsenite-induced cell transformation 4. In this study, we further demonstrated that m6A mediated arsenite-induced cell transformation by inhibiting p53 activation rather than by altering p53 expression (Fig. 3–4). However, our experiments have several limitations. First, in our studies, the wound-healing assay was used to determine cell invasion. The results from the assay may not completely match the results from the cell-transformation experiments. Second, since we used transient transfection of short interfering RNA (siRNAs) to knockdown genes, we could not detect the effects of siRNA on HaCaT cells using the soft agar clone formation assay and plate colony formation assay. This is because both assays require the growth of cells for at least 10 days. Thus, we only detected the effects of transient siRNA transfection on the transformation of HaCaT cells using cell viability assay, wound-healing assay, and cell apoptosis, which only require short period of time (24 h).
In this study, we tested p53 activation by determining p53 activity, p53 phosphorylation and acetylation, p53 localization, the expression of p53 target genes and p53 regulators, and apoptosis to demonstrate p53 activation. p53 activity was determined based on the principle that p53 protein binds to a consensus p53-binding site and activates the expression of downstream genes 21,22 using a commercial p53 Activity Assay kit. However, it should be noted that the activation of p53 and the regulation of its downstream targets are also mediated by p53 posttranslational modifications and modulated by p53 regulators 39. Our approaches not only have demonstrated the p53 activation from different aspects but also are beneficial for the future research on understanding the mechanisms of p53 inactivation mediated by m6A in arsenite-induced cell transformation.
There has been a controversy regarding whether chronic arsenite exposure can lead to carcinogenesis by inducing p53 mutations based on population-based studies 40–42. This is because the results from the population-based studies are usually affected by a variety of confounding factors, which can introduce the bias to the results of p53 mutational studies, especially those from the human population that was chronically exposed to low doses of arsenite. In addition, the population-based analysis has a limitation for detecting precise alterations of p53 in cells. Using cell models, we were able to show that chronic arsenite exposure at low doses did not induce the p53 gene mutations in its exons 5–9 19,438(Fig. 3a). The results are consistent with a report showing that no p53 mutations in exons 5–8 are detected in rats that are exposed to the metabolite of arsenite, dimethylarsinic acid 44 Thus, our results suggest that it is unlikely that p53 mutations and aberrant expression were involved in arsenite-induced human keratinocytes transformation that was mediated by m6A. Instead, it was the suppression of p53 activation that mediated arsenite-induced human keratinocyte transformation and carcinogenesis. This is also supported by the studies that have shown that arsenite suppresses p53 activation by decreasing p53 acetylation and phosphorylation13, inducing p53 poly(ADP-ribosyl)ation 45, and inhibiting the DNA binding activity of p53 and its target gene p21 expression 46. Furthermore, it has been found that microRNA, miR-410, −548ac, and −548a-3p, and p53 regulators, HMGB1 and MDM2 can also be involved in the suppression of p53 activation induced by arsenite in cellular transformation and carcinogenesis 13,47. However, our results cannot exclude a possibility that m6A transcripts in p53 may facilitate the selection of a mutated codon in the other encoding regions of the protein through m6A in TP53 codon RNA transcripts that promote the selection of m6A-R273H pre-mRNA for splicing producing the mutant protein in cancer cells 48. Thus, our study revealed the m6A-mediated mechanism underlying arsenite-induced human keratinocyte transformation, shedding light on the complex mechanisms by which arsenite induces alterations of p53 activation in cell transformation and carcinogenesis.
It is suggested that there is a significant association between cutaneous melanoma and exposure to arsenical pesticides 49. It is also found that the mortality rate of melanoma is significantly increased in the population that was occupationally exposed to arsenic 50. Moreover, the increased mortality of skin cancer, including melanoma is higher than the predicted rate resulting from aging suggesting that the increased mortality results from environmental exposure such as arsenic exposure 51. Further, since both melanocytes and keratinocytes are target cells in the skin and share the same mechanism by which arsenic inhibits DNA repair, chronic arsenic exposure may promote the accumulation of unrepaired DNA damage in melanocytes causing melanoma 52. Thus, all these studies point to a strong correlation between skin cutaneous melanoma (SKCM) and arsenic exposure supporting a notion that arsenite exposure is a risk factor of SKCM. Our bioinformatic analysis of SKCM from several human studies further demonstrated that m6A methyltransferases and readers were associated with the development of SKCM, suggesting an important role of m6A in mediating the development of skin cancer. Our bioinformatics analysis of p53 mutation and expression, its target genes, and regulators in SKCM further indicated that p53 inactivation played a crucial role in the development of skin cancer. Our bioinformatics analysis of the co-expression of both m6A methyltransferases and p53 regulators in SKCM tissue also indicated a correlation between m6A and p53 in skin cancer. Thus, our bioinformatics analysis results support the conclusions from our experimental results.
Since its discovery in 2012 8, the cellular function of m6A and its regulation have been studied extensively, and its “writer”, “eraser”, and “reader” have been identified, and its roles in various biological processes and the development of diseases have been reported 17,53. Recent studies have also demonstrated that m6A serves as a beacon for the recruitment of a repair DNA polymerase to UV damage sites 54 and is involved in neurite outgrowth recovery in mouse neurons that are exposed to arsenite 55 indicating its important role in the arsenite-driven hormesis on cytotoxicity 5. Further, our bioinformatic analysis on the network of m6A- and p53-related genes in this study demonstrated that one function of the m6A- and p53-related genes in the network is to mediate cellular response to hypoxia and oxygen levels (Supplementary Fig. 6c). This is also consistent with the other study 55 that suggests that there is an association between m6A and oxidative stress. This is further supported by our recent findings showing that m6A is associated with oxidative stress in arsenite-driven hormesis 5, and arsenite-induced oxidative stress increases the levels of m6A 6. All these studies suggest the critical roles of m6A in mediating and modulating geno- and cytotoxicity of environmental toxicants including arsenite. The roles of m6A in oxidative stress, as well as the toxic effects of other environmental toxicants and their underlying mechanisms, need to be further elucidated in the future.
Supplementary Material
Highlights.
The level of m6A is increased in arsenite-transformed human keratinocytes
m6A mediates migration and apoptosis of arsenite-transformed keratinocytes
m6A attenuates p53 activation instead of expression in arsenite-transformed cells
m6A suppresses p53 by altering the expression of p53 activation regulators
m6A writers, erasers, and readers mediates the function of m6A on p53 inactivation
Acknowledgments
We appreciate the technical support of the Public Health and Preventive Medicine Provincial Experiment Teaching Center at Sichuan University and Food Safety Monitoring and Risk Assessment Key Laboratory of Sichuan Province.
Funding
Funding: This work was supported by the grant from the National Science Foundation of China (No. 81773380). Y. Lai and Y. Liu are supported by NIHR01ES023569.
Abbreviations
- m6A
N6-methyladenosine
- IARC
International Agency for Research on Cancer
- PBS
hosphate-buffered saline
- FITC
fluorescein isothiocyanate
- PI
propidium iodide
- DAPI
4′,6-diamidino-2-phenylindole
- TCGA
The Cancer Genome Atlas
- SKCM
skin cutaneous melanoma
- SEM
standard error of the mean
- METTL3
methyltransferase-like 3
- METTL14
methyltransferase-like 14
- WTAP
Wilms tumor 1-associated protein
- VIRMA/KIAA1429
via like m6A methyltransferase associated
- FTO
fat mass and obesity-associated protein
- YTHDF1
YTH N6-methyladenosine RNA binding protein 1
- YTHDF1
YTH N6-methyladenosine RNA binding protein 2
- CDKN1A
cyclin dependent kinase inhibitor 1A
- RRM2B
ribonucleotide reductase regulatory TP53 inducible subunit M2B
- SUSD6/ KIAA0247
sushi domain containing 6
- PRDM2
PR/SET domain 2
- MDM2
murine double minute 2
- YY1
Yin Yang-1
Footnotes
Competing interests
The authors have declared that no competing interests exist.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sen P, Biswas T Arsenic: the largest mass poisoning of a population in history. BMJ 346, f3625, doi: 10.1136/bmj.f3625 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Navas-Acien A, Nachman KE. Public health responses to arsenic in rice and other foods. JAMA Intern Med 173(15), 1395–1396, doi: 10.1001/jamainternmed.2013.6405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Humans, I. W. G. o. t. E. o. C. R. t. Arsenic, metals, fibres, and dusts. IARC Monogr Eval Carcinog Risks Hum 100, 11–465 (2012). [PMC free article] [PubMed] [Google Scholar]
- 4.Gu S, Sun D, Dai H & Zhang Z N6-methyladenosine mediates the cellular proliferation and apoptosis via microRNAs in arsenite-transformed cells. Toxicol Lett 292, 1–11, doi: 10.1016/j.toxlet.2018.04.018 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Zhao T, Sun D, Wu M & Zhang Z Changes of RNA N6-methyladenosine in the hormesis effect induced by arsenite on human keratinocyte cells. Toxicol In Vitro 56, 84–92, doi: 10.1016/j.tiv.2019.01.010 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Zhao T, Li X, Sun D & Zhang Z Oxidative stress: One potential factor for arsenite-induced increase of N6-methyladenosine in human keratinocytes. Environ Toxicol Pharmacol 69, 95–103, doi: 10.1016/j.etap.2019.04.005 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Bai L et al. m6A demethylase FTO regulates dopaminergic neurotransmission deficits caused by arsenite. Toxicol Sci, doi: 10.1093/toxsci/kfy172 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Dominissini D et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206, doi: 10.1038/nature11112 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Kwok CT, Marshall AD, Rasko JE & Wong JJ Genetic alterations of m6A regulators predict poorer survival in acute myeloid leukemia. J Hematol Oncol 10, 39–45, doi: 10.1186/s13045-017-0410-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng G et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49, 18–29, doi: 10.1016/j.molcel.2012.10.015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y et al. Induction of cytoplasmic accumulation of p53: a mechanism for low levels of arsenic exposure to predispose cells for malignant transformation. Cancer Res 68, 9131–9136, doi: 10.1158/0008-5472.CAN-08-3025 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y et al. Low dose arsenite confers resistance to UV induced apoptosis via p53-MDM2 pathway in ketatinocytes. Oncogenesis 6, e370, doi: 10.1038/oncsis.2017.67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Eryani L et al. Differentially Expressed mRNA Targets of Differentially Expressed miRNAs Predict Changes in the TP53 Axis and Carcinogenesis-Related Pathways in Human Keratinocytes Chronically Exposed to Arsenic. Toxicol Sci 162, 645–654, doi: 10.1093/toxsci/kfx292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Y et al. m6AVar: a database of functional variants involved in m6A modification. Nucleic Acids Res 46, D139–D145, doi: 10.1093/nar/gkx895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shannon P et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504, doi: 10.1101/gr.1239303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu Y et al. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics 11, 8–17, doi: 10.1016/j.gpb.2012.12.002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bi Z et al. A dynamic reversible RNA N6 -methyladenosine modification: current status and perspectives. J Cell Physiol, doi: 10.1002/jcp.28014 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Pan Y, Ma P, Liu Y, Li W & Shu Y Multiple functions of m6A RNA methylation in cancer. J Hematol Oncol 11, 48, doi: 10.1186/s13045-018-0590-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leroy B, Anderson M & Soussi T TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat 35, 672–688, doi: 10.1002/humu.22552 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto S & Iwakuma T Regulators of Oncogenic Mutant TP53 Gain of Function. Cancers (Basel) 11, doi: 10.3390/cancers11010004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raj N & Attardi LD The Transactivation Domains of the p53 Protein. Cold Spring Harb Perspect Med 7, doi: 10.1101/cshperspect.a026047 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saha T, Kar RK & Sa G Structural and sequential context of p53: A review of experimental and theoretical evidence. Prog Biophys Mol Biol 117, 250–263, doi: 10.1016/j.pbiomolbio.2014.12.002 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Georgakilas AG, Martin OA & Bonner WM p21: A Two-Faced Genome Guardian. Trends Mol Med 23, 310–319, doi: 10.1016/j.molmed.2017.02.001 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Pitceathly RD. et al. Adults with RRM2B-related mitochondrial disease have distinct clinical and molecular characteristics. Brain 135, 3392–3403, doi: 10.1093/brain/aws231. Epub 2012 Oct 29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polato F et al. DRAGO (KIAA0247), a new DNA damage-responsive, p53-inducible gene that cooperates with p53 as oncosuppressor. [Corrected]. J Natl Cancer Inst 106, dju053, doi: 10.1093/jnci/dju053 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan F et al. Up-regulation of Siah1 by ethanol triggers apoptosis in neural crest cells through p38 MAPK-mediated activation of p53 signaling pathway. Arch Toxicol 91, 775–784, doi: 10.1007/s00204-016-1746-3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lagunas-Martinez A et al. MG132 plus apoptosis antigen-1 (APO-1) antibody cooperate to restore p53 activity inducing autophagy and p53-dependent apoptosis in HPV16 E6-expressing keratinocytes. Apoptosis 22, 27–40, doi: 10.1007/s10495-016-1299-1 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Ishak Gabra MB et al. IKKβ activates p53 to promote cancer cell adaptation to glutamine deprivation. Oncogenesis 7, 93, doi: 10.1038/s41389-018-0104-0 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu CE et al. Targeting negative regulation of p53 by MDM2 and WIP1 as a therapeutic strategy in cutaneous melanoma. Br J Cancer 118, 495–508, doi: 10.1038/bjc.2017.433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shadat NM et al. Retinoblastoma protein-interacting zinc finger 1 (RIZ1) regulates the proliferation of monocytic leukemia cells via activation of p53. Cancer Invest 28, 806–812, doi: 10.3109/07357907.2010.494323 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Tackmann NR & Zhang Y Mouse modelling of the MDM2/MDMX-p53 signalling axis. J Mol Cell Biol 9, 34–44, doi: 10.1093/jmcb/mjx006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue K, Fry EA & Frazier DP Transcription factors that interact with p53 and Mdm2. Int J Cancer 138, 1577–1585, doi: 10.1002/ijc.29663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei J et al. Escape, or Vanish: Control the Fate of p53 through MDM2-Mediated Ubiquitination. Anticancer Agents Med Chem 16, 174–189, doi: 10.2174/1871520615666150907093358 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Agarwala SD. et al. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet 8, e1002732, doi: 10.1371/journal.pgen.1002732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geula S et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006, doi: 10.1126/science.1261417 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Zhong S et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288, doi: 10.1105/tpc.108.058883 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vu LP. et al. The Biology of m6A RNA Methylation in Normal and Malignant Hematopoiesis. Cancer Discov 9(1), 25–33, doi: 10.1158/2159-8290.CD-18-0959 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Wei CM. et al. Methylated nucleotides block 5′terminus of HeLa cell messenger RNA. Cell 4(4), 379–386, doi: 10.1016/0092-8674(75)90158-0 (1975). [DOI] [PubMed] [Google Scholar]
- 39.Riley T et al. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9(5), 402–412, doi: 10.1038/nrm2395 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Kelsey KT et al. TP53 alterations and patterns of carcinogen exposure in a U.S. population-based study of bladder cancer. Int J Cancer 117, 370–375, doi: 10.1002/ijc.21195 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Zhang A et al. Unventilated indoor coal-fired stoves in Guizhou province, China: cellular and genetic damage in villagers exposed to arsenic in food and air. Environ Health Perspect 115, 653–658, doi: 10.1289/ehp.9272 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore LE et al. P53 alterations in bladder tumors from arsenic and tobacco exposed patients. Carcinogenesis 24, 1785–1791, doi: 10.1093/carcin/bgg136 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Bykov VJN, Eriksson SE, Bianchi J & Wiman KG Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer 18, 89–102, doi: 10.1038/nrc.2017.109 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Wei M et al. Carcinogenicity of dimethylarsinic acid in male F344 rats and genetic alterations in induced urinary bladder tumors. Carcinogenesis 23, 1387–1397, doi: 10.1093/carcin/23.8.1387 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Komissarova EV & Rossman TG Arsenite induced poly(ADP-ribosyl)ation of tumor suppressor P53 in human skin keratinocytes as a possible mechanism for carcinogenesis associated with arsenic exposure. Toxicol Appl Pharmacol 243, 399–404, doi: 10.1016/j.taap.2009.12.014 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang F et al. Arsenite inhibits p53 phosphorylation, DNA binding activity, and p53 target gene p21 expression in mouse epidermal JB6 cells. Mol Carcinog 45, 861–870, doi: 10.1002/mc.20245 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Pant V & Lozano G Dissecting the p53-Mdm2 feedback loop in vivo: uncoupling the role in p53 stability and activity. Oncotarget 5, 1149–1156, doi: 10.18632/oncotarget.1797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uddin MB. et al. An N6-methyladenosine at the transited codon 273 of p53 pre-mRNA promotes the expression of R273H mutant protein and drug resistance of cancer cells. Biochem Pharmacol 160, 134–145, doi: 10.1016/j.bcp.2018.12.014. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dennis LK. et al. Pesticide use and cutaneous melanoma in pesticide applicators in the agricultural heath study. Environ Health Perspect 118(6), 812–817, doi: 10.1289/ehp.0901518. (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gianicolo EAL. et al. Long-term effect of arsenic exposure: Results from an occupational cohort study. Am J Ind Med 62(2), 145–155, doi: 10.1002/ajim.22939. (2019). [DOI] [PubMed] [Google Scholar]
- 51.Alonso FT. et al. Increased skin cancer mortality in Chile beyond the effect of ageing: Temporal analysis 1990 to 2005. Acta Derm Venereol 90(2), 141–146, doi: 10.2340/00015555-0787. (2010). [DOI] [PubMed] [Google Scholar]
- 52.Cooper KL. et al. Melanocytes and keratinocytes have distinct and shared responses to ultraviolet radiation and arsenic. Toxicol Lett 224(3), 407–415, doi: 10.1016/j.toxlet.2013.11.010. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maity A & Das B N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases. FEBS J 283, 1607–1630, doi: 10.1111/febs.13614 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Xiang Y et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576, doi: 10.1038/nature21671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li M et al. Ythdf2-mediated m6A mRNA clearance modulates neural development in mice. Genome Biol 19, 69–85, doi: 10.1186/s13059-018-1436-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source data for Figs. 1–6 are provided in Supplementary Table 1–5. The source data for Supplementary Figs. 1–6 are provided in Supplementary Table 6. The source data for p53 sequence are provided in the Supplementary p53 sequence.
Fig. 1. m6A level increases in arsenite-transformed human keratinocytes.
a, Cell proliferation of HaCaT and HaCaT-T cells was measured by MTT assay. b, Percentage of clone formation was calculated by soft agar clone formation assay in HaCaT and HaCaT-T cells (down). Representative images of HaCaT and HaCaT-T cells in soft agar clone formation assay observed with a microscope at 400× magnification (up). c, Percentage of colony formation was tested by plate colony formation assay in HaCaT and HaCaT-T cells (down). Representative images of HaCaT and HaCaT-T cells in plate colony formation (up). d, Total m6A levels in HaCaT and HaCaT-T cells of 0, 15, 30, 50 passages were determined by EpiQuik m6A RNA Methylation Quantification Kit. e, f, RT-qPCR testing the mRNA levels (e) and western blotting examining the protein levels (f) of m6A regulating proteins (METTL3, METTL14, WTAP, KIAA1429, FTO, and YTHDF1) in HaCaT and HaCaT-T cells of 0, 15, 30, 50 passages are shown in heat maps. Representative images of immunoblot (f, left). For a-f, n = at least 3 biological replicates, error bars indicate mean ± SEM. The P-values were determined by two-tailed t-test. “*” indicates a significant difference compared to control group (P < 0.05).
Fig. 6. m6A methylation inhibits PRDM2 expression and increases MDM2 and YY1 levels in HaCaT-T cells.
a-d, m6A levels in p53 (a), PRDM2 (b), MDM2 (c), and YY1 (d) mRNA of HaCaT and HaCaT-T cells were determined by MeRIP. e, The mRNA levels of PRDM2, MDM2, and YY1 in HaCaT-T cells with introduction of YTHDF1 siRNA (siYTHDF1) or YTHDF2 siRNA (siYTHDF2) were tested by RT-qPCR. f, The representative images of MDM2 (left) and YY1 (middle) in HaCaT-T cells with introduction of two YTHDF1 siRNA (siYTHDF1–1 and siYTHDF1–2). Quantitative protein levels of MDM2 and YY1 in HaCaT-T cells with introduction of two YTHDF1 siRNA were detected by western blotting (right). g, The representative images (left) of PRDM2 in HaCaT-T cells with introduction of two YTHDF2 siRNA (siYTHDF2–1 and siYTHDF2–2). Quantitative protein levels of PRDM2 in HaCaT-T cells with introduction of two YTHDF2 siRNA were detected by western blotting (right). For a-g, n = 3 biological replicates, error bars indicate mean ± SEM. The P-values were determined by two-tailed t-test. “*” indicates a significant difference compared to control group (P < 0.05).