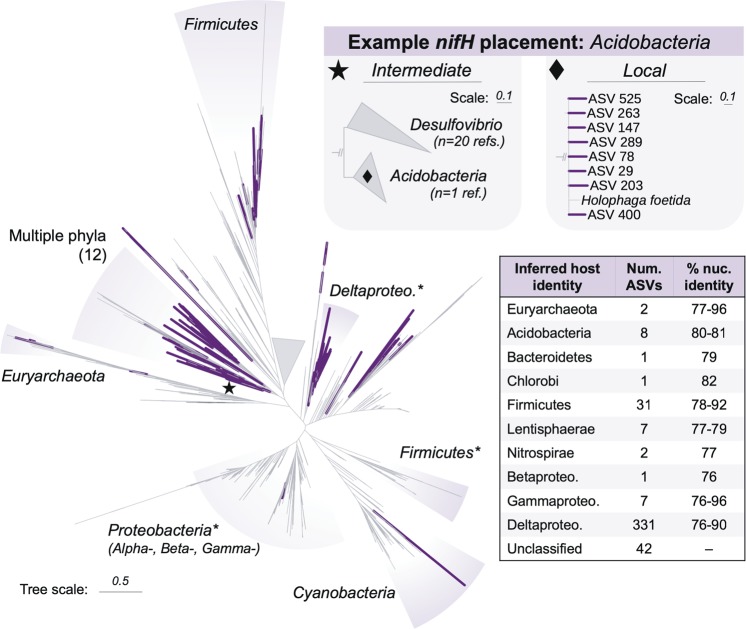
Fig. 4. Diversity of recovered nifH sequences.
Bold branches show nifH ASVs (n = 434) placed at their maximum likelihood positions on the reference nifH tree (n = 6040). Shaded regions indicate taxonomic identities of select clades for reference. Gray wedge shows collapsed branches of nifH homologs; the full tree including these sequences can be found in Supplementary Fig. S7. Reference sequences on terminal branches with lengths >1.0 were pruned for clarity (n = 34). Phyla within clade labeled “Multiple phyla”: Acidobacteria, Actinobacteria, Bacteroidetes, Chlorobi, Chloroflexi, Cyanobacteria, Elusimicrobia, Lentisphaerae, Nitrospirae, Proteobacteria, Spirochaetes, and Verrucomicrobia. Star and diamond insets display example nifH ASV placements at finer phylogenetic scales. ‘% nuc. identity’ in table shows range of percent nucleotide identities between nifH ASVs and closest references for each taxonomic group. *clades which include one or two nifH sequences from other phyla (clades without asterisks contain sequences from one phylum).