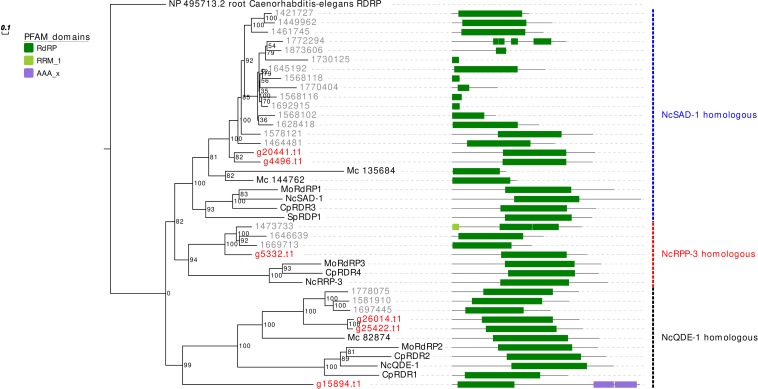
FIGURE 2.
Phylogenetic analysis and PFAM domains of RdRp proteins. Sequences are discernible by species according to a two-letter prefix and/or a color code: Mo, Magnaporthe oryzae; Nc, Neurospora crassa, Mc, Mucor circinelloides; Sp, Schizosaccharomyces pombe; Cp, Cryphonectria parasitica; gray, Rhizophagus irregularis; red, Gigaspora margarita. Protein ID (NCBI or JGI): MoRdRP1 = XP_003721007.1, MoRdRP2 = XP_003711624.1, MoRdRP3 = XP_003712093.1, NcQDE-1 = EAA29811.1, NcSAD-1 = XP_964248.3, NcRRP-3 = XP_963405.1, SpRDP1 = NP_001342838.1, McRdRP-1 = 111871, McRdRP-2 = 104159, CpRDR1 = 270014, CpRDR2 = 35624, CpRDR3 = 10929, CpRDR4 = 339656. R. irregularis proteins are identified by JGI numeric codes. G. margarita proteins are identified by Venice et al. (2019) annotation code. The numbers at the nodes are bootstrap values (%) for 1000 replications. Tree was rooted using Caenorhabditis elegans RdRP (NCBI Reference Sequence: NP_495713.2). Tree was reshaped on root with Newick Utilities v1.6 and visualized with Evolview v3.