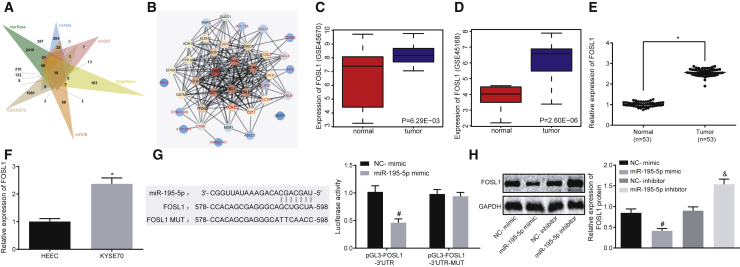
Figure 3.
As a Target Gene of miR-195-5p, FOSL1 Is Highly Expressed in EC
(A) Intersection of the target genes of miR-195-5p predicted by StarBase, DIANA, mirDIP, TargetScan, and miRDB and DEGs in the GEO: GSE45670 dataset. (B) PPI network of top 20 EC-related genes from DisGeNET and the 18 intersected DEGs from panel, in which red characters indicate DEGs that bind to miR-195-5p, black characters indicate EC-related genes, and circle color indicates the correlation degree with other genes where red indicates close correlation. (C) Relative expression of FOSL1 in EC and normal samples based on the GEO: GSE45670 dataset. (D) Relative expression of FOSL1 in EC and normal samples based on the GEO: GSE45168 dataset. (E) Expression of FOSL1 in EC tissues and adjacent normal tissues (n = 53) determined by qRT-PCR. (F) Expression of FOSL1 in EC cell line KYSE70 and HEECs determined by qRT-PCR. (G) Relationship between miR-195-5p and FOSL1 analyzed by detecting the luciferase activity of pGL3-FOSL1-3′ UTR and pGL3-FOSL1-3′ UTR-MUT after transfection with miR-195-5p mimic or NC-mimic through dual-luciferase reporter assay. (H) Expression of FOSL1 in KYSE70 cells transfected with miR-195-5p mimic or miR-195-5p inhibitor measured by western blot analysis. *p < 0.05 compared with adjacent normal tissues or HEECs; #p < 0.05 compared with KYSE70 cells transfected with NC-mimic; &p < 0.05 compared with KYSE70 cells transfected with NC-inhibitor. The measurement data are expressed as mean ± standard deviation. Data in (C)–(E) were compared by paired t test. Data in (F)–(H) were compared by unpaired Student’s t test. Data among multiple groups were compared by one-way ANOVA and Tukey’s post hoc test. The experiment was repeated three times.