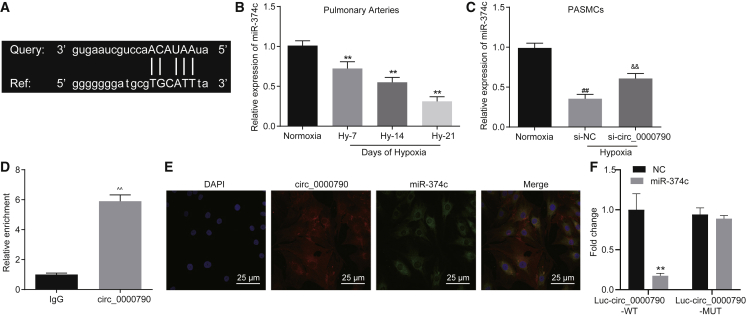
Figure 4.
mmu_circ_0000790 Binds to miR-374c
(A) Bioinformatics prediction of sequence containing the miR-374c binding site in mmu_circ_0000790 3′ UTR. (B) Expression of miR-374c in pulmonary artery of normoxia and hypoxia mice detected by qRT-PCR. (C) Expression of miR-374c in hypoxic PASMCs detected by qRT-PCR. (D) Enrichment of mmu_circ_0000790-specific probes detected by RIP assay. (E) Cellular localization of mmu_circ_0000790 detected by a FISH assay. (F) The binding of circ_0000790 to miR-374c confirmed by a dual-luciferase reporter gene assay. **p < 0.01 versus the normoxia mice; ##p < 0.01 versus normoxia cells; &&p < 0.01 versus the si-NC; ˆˆp < 0.01 versus the IgG group. N = 5. The data (mean ± standard deviation) between two groups were compared using a t test, and those among multiple groups were compared with one-way ANOVA. Each cell experiment was run in triplicate independently. miR-374c, microRNA-374c; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction; HPH, hypoxic pulmonary hypertension; PASMC, pulmonary artery smooth muscle cell; RIP, RNA-binding protein immunoprecipitation; FISH, fluorescent in situ hybridization; NC negative control; ANOVA, analysis of variance.