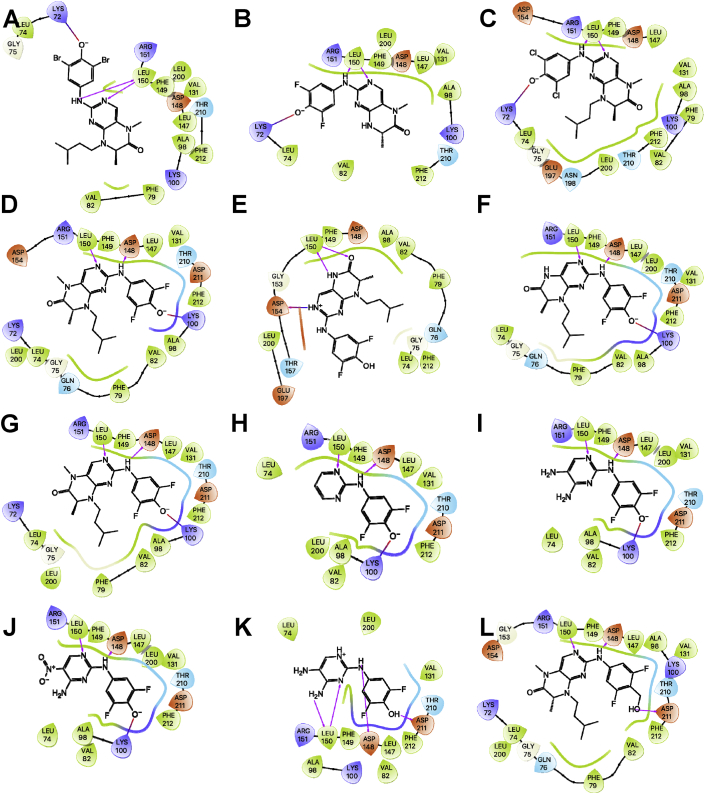
Fig. 6.
Ligand interaction map of the predicted binding mode of A) 32R-isomer deprotonated, B) 39S-isomer deprotonated, C) 31R-isomer deprotonated, D) BI-D1870 R-isomer deprotonated, E) 33S-isomer protonated, F) 33S-isomer deprotonated, G) BI-D1870 S-isomer deprotonated, H) 38 deprotonated, I) 44 deprotonated, J) 43 deprotonated, K) 44 protonated, and L) 26R-isomer in the ATP-binding site of the RSK2 NTKD, where red residues are charged negative, purple residues are charged positive, green residues are hydrophobic, and blue residues are polar, magenta arrows indicate H-bonds, violet lines indicate slat bridges, and gray spheres represent areas of solvent exposure.