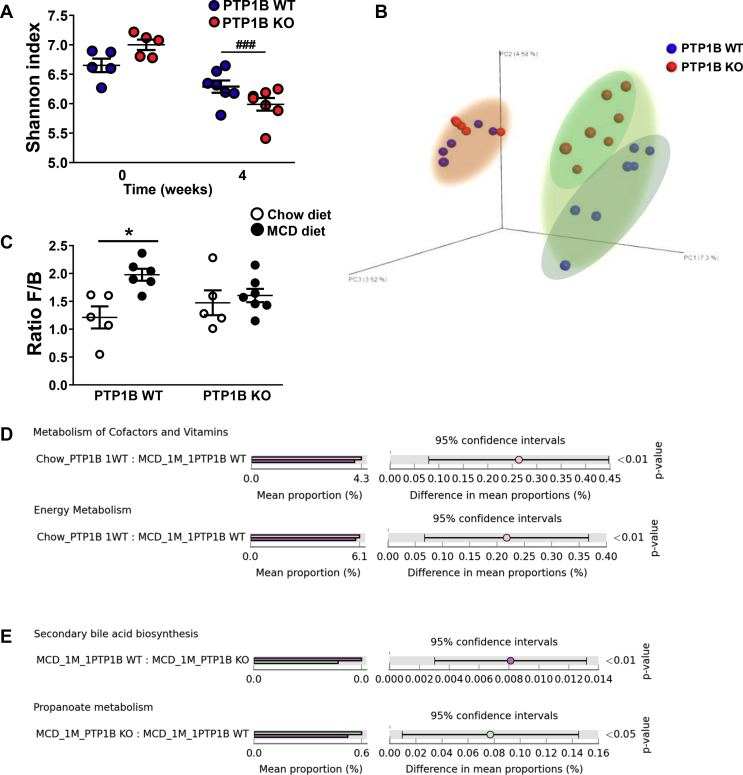
Figure 1.
Gut microbiota composition and functional changes. A) Alpha-diversity analysis for PTP1B WT and PTP1B KO mice fed a chow or MCD diet for 4 weeks. Shannon index is expressed as mean ± SEM. ###p < 0.001 PTP1B KO chow vs PTP1B KO MCD. B) Phylogenetic β-diversity of fecal microbiota. Graph represents principal coordinate analysis (PCoA) plot based on 16 S rRNA gene sequencing of stool samples from PTP1B WT (blue symbols) and PTP1B KO (red symbols) mice fed a chow or MCD diet for 4 weeks. Differences between genotypes in this case are indicated with different clouds. Pink cloud corresponds to animals fed chow diet and green cloud surrounds animals fed a MCD diet. C) Ratio Firmicutes/Bacteroidetes (F/B) is expressed as mean ± SEM. ∗p < 0.05 PTP1B WT chow vs PTP1B WT MCD. D-E) Predicted functional changes in gut microbiota analyzed by PICRUSt tool. Bar graphs show significant differences in KEGG pathways. MCD_1 M refers to animals fed a MCD diet for 4 weeks; Chow_ refers to animals fed a chow diet. Significant differences are shown as p-value at the right panel. n = 5–7 mice per experimental group.