Early-childhood caries is one of the most prevalent diseases in children worldwide and, while preventable, remains a global public health concern. Untreated cavities are painful and expensive and can lead to tooth loss and a lower quality of life. Caries are driven by acid production via microbial fermentation of dietary carbohydrates, resulting in enamel erosion. While caries is a well-studied disease, most research has focused on bacterial impacts, even though fungi are commensal organisms living within the plaque biofilm. There is very little known about how fungi impact caries pathogenicity. The elucidation of fungal taxa involved in caries disease progression is necessary for a more holistic view of the human oral microbiome. Data from this study will improve our understanding of how the fungal community changes as disease progresses and provide insight into the complex etiology of dental caries, which is necessary for the development of treatment plans and preventative measures.
KEYWORDS: dental caries, mycobiome, ITS, amplicon, microbiome, fungi, oral, oral microbiology
ABSTRACT
Dental caries is one of the most common diseases worldwide. Bacteria and fungi are both commensals in the oral cavity; however, most research regarding caries has focused on bacterial impacts. The oral fungal mycobiome associated with caries is not well characterized, and its role in disease is unclear. ITS1 amplicon sequencing was used to generate taxonomic profiles from site-specific supragingival plaque samples (n = 82) obtained from 33 children with different caries status. Children were either caries free (CF), caries active with enamel lesions (CAE), or caries active with dentin lesions (CA). Plaque samples were collected from caries-free surfaces (PF) and from enamel (PE) and dentin (PD) lesions. Taxonomic profiles representing the different categorizations (CF-PF, CAE-PF, CAE-PE, CA-PF, CA-PE, and CA-PD) were used to characterize the mycobiome and its change through disease progression. A total of 139 fungal species were identified. Candida albicans was the most abundant species, followed by Candida dubliniensis. We found that severely progressed plaque communities (CA-PD) were significantly different from healthy plaque communities (CF-PF). A total of 32 taxa were differentially abundant across the plaque categories. C. albicans, C. dubliniensis, Nigrospora oryzae, and an unclassified Microdochium sp. were correlated with caries, whereas 12 other taxa were correlated with health. C. dubliniensis increased steadily as caries progressed, suggesting that C. dubliniensis may play an important role in caries pathogenicity. In contrast, four health-associated fungal taxa have the potential to antagonize the cariogen Streptococcus mutans via xylitol production, suggesting a possible fungal mechanism that could contribute to maintenance of dental health.
IMPORTANCE Early-childhood caries is one of the most prevalent diseases in children worldwide and, while preventable, remains a global public health concern. Untreated cavities are painful and expensive and can lead to tooth loss and a lower quality of life. Caries are driven by acid production via microbial fermentation of dietary carbohydrates, resulting in enamel erosion. While caries is a well-studied disease, most research has focused on bacterial impacts, even though fungi are commensal organisms living within the plaque biofilm. There is very little known about how fungi impact caries pathogenicity. The elucidation of fungal taxa involved in caries disease progression is necessary for a more holistic view of the human oral microbiome. Data from this study will improve our understanding of how the fungal community changes as disease progresses and provide insight into the complex etiology of dental caries, which is necessary for the development of treatment plans and preventative measures.
INTRODUCTION
Early-childhood caries (ECC) is one of the most prevalent diseases in children, and, while preventable, it remains a global public health problem (1–3). The incidence of ECC in the United States is estimated between 3 and 6%, and in less developed countries, ECC continues to grow rapidly, with an incidence of up to 70% (3). Often, ECC remains untreated and leads to adverse effects on the child’s overall health, development, and quality of life (3). Due to the significant detrimental effects of ECC on quality of life and the substantial economic burden associated with this disease, preventative measures and early treatment are crucial.
Dental caries is a complex, multifactorial, polymicrobial, and biofilm-induced disease resulting in loss of tooth hard tissues. Caries is driven by acid production via bacterial metabolism of easily fermentable carbohydrates such as sucrose (4, 5). Despite the wealth of studies on ECC and caries, the ecological aspects of the disease are still not fully understood. The focus of many studies examining the microbiology of caries has been on bacterial members of the tooth microbiome (6–9). More recently, studies have highlighted the presence of fungi in the oral cavity and in supragingival plaque of individuals presenting either oral health, caries, or other oral diseases (10–18).
The oral fungal community is not well characterized; this is particularly true regarding caries. The development of next-generation sequencing (NGS) technologies has provided an affordable method to identify and quantify members of the oral mycobiome. The internally transcribed spacer (ITS) 1 and 2 regions have been widely used for amplicon sequencing of fungal communities due to their ability to distinguish between a wide array of fungal taxa (19). A study from 2010 used the ITS1 region to classify the health-associated oral fungal mycobiome and highlighted the vast diversity of fungal taxa in the mouth (10). Since then, over 150 fungal species have been identified in the oral cavity using the ITS1 and ITS2 regions. In almost all of these studies, Candida spp., particularly Candida albicans, have dominated the composition of the oral mycobiome (10, 13, 16–18). Evidence of the role that C. albicans plays in caries has been contradictory. Some studies have observed no significant differences between caries status (caries free or caries active) and C. albicans prevalence (20, 21), while another showed there was no correlation between caries risk and C. albicans presence (22). In contrast, a study using a rodent caries model found that C. albicans had the ability to cause caries when fed a high-sucrose and -glucose diet (23). Moreover, other studies have found that C. albicans is commonly detected in association with the caries pathogen Streptococcus mutans in plaque samples from children with ECC (24–26). In one of these studies, increased glucosyltransferase activity of S. mutans (a caries virulence factor) was significantly elevated in the presence of C. albicans, highlighting an important cross-kingdom association (26). In vitro testing with Candida sp. has shown increased adherence of bacterial organisms on teeth when Candida species are present. It has been proposed that the Candida species act as a bridge to which oral bacteria can adhere, providing more surface area for extracellular polysaccharide (EPS) formation while also making bacteria less susceptible to antibiotics and antimicrobials (27). Oral bacterial and fungal interactions may be important risk factors and indicators in caries disease (27–29). To begin to elucidate the complex microbial interactions contributing to caries, a better characterization of the mycobiome of supragingival dental plaque during the stages of caries progression is needed.
This study used high-throughput amplicon sequencing of the ITS1 region to characterize the oral mycobiome associated with dental health and ECC and is only the second study to characterize the oral mycobiome associated with caries. Furthermore, we used a site-specific sampling approach to measure changes in mycobiome composition through the progressive stages of the disease. We hypothesized that the community composition would change as disease progressed. A characterization of the fungal community as caries progresses will allow for further investigation into the complex interactions between the bacterial and fungal microbiomes and how these interactions impact dental health and caries disease development and progression.
RESULTS
Illumina sequencing.
The total number of reads after Illumina MiSeq sequencing was 14,448,675, with an average number of reads per sample of 138,840. After completing an initial quality check (QC) on the samples to remove low-quality reads, chimeric sequences, and PhiX reads, the total number of reads was reduced to 4,946,479. The average number of reads per sample was 67,759, with a minimum of 2,194 and a maximum of 168,228. Ten samples with low-quality reads, low numbers of reads, or low operational taxonomic unit (OTU) counts were removed from the analysis. A total of 412 taxonomic assignments were generated. OTUs with the same taxonomic classification were collapsed, and singletons were removed, resulting in a total of 214 unique taxonomic assignments composed of 2 phyla, 184 genera, and 139 species.
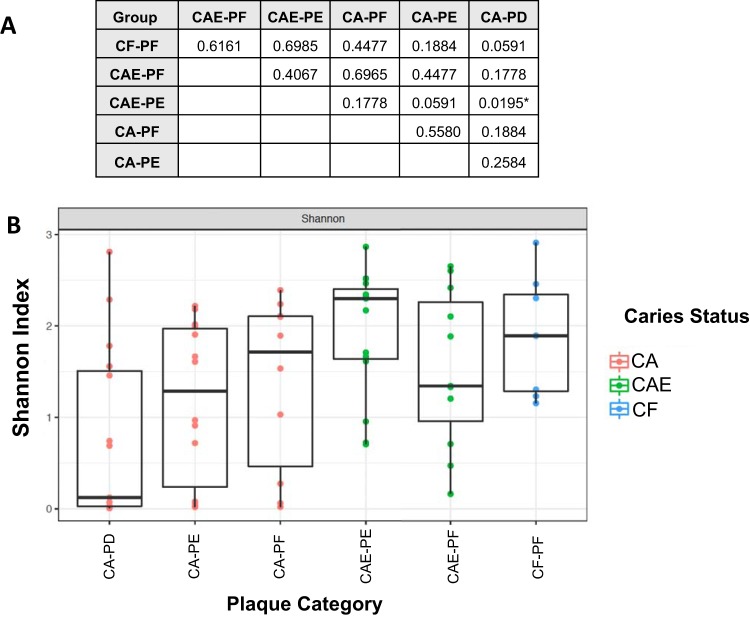
Children were either caries free (CF), caries active with enamel lesions (CAE), or caries active with dentin lesions (CA). Plaque samples were collected from caries-free surfaces (PF) and from enamel (PE) and dentin (PD) lesions. Significant differences in species richness were only observed between CAE-PE and CA-PD communities (P = 0.019) (Fig. 1A). Overall, fungal community richness decreased as caries progressed, with the highest alpha diversity observed in CF-PF communities and lowest alpha diversity observed in CA-PD communities (Fig. 1B).
FIG 1.
Shannon alpha diversity metrics of site-specific dental plaque mycobiome samples as dental caries progresses. (A) Pairwise Kruskal-Wallis test results. P values were corrected for multiple comparisons using FDR correction. *, statistical significance (α < 0.05). (B) Box plot depicting Shannon alpha diversity measures for each sample category. Colors represent caries status of the child.
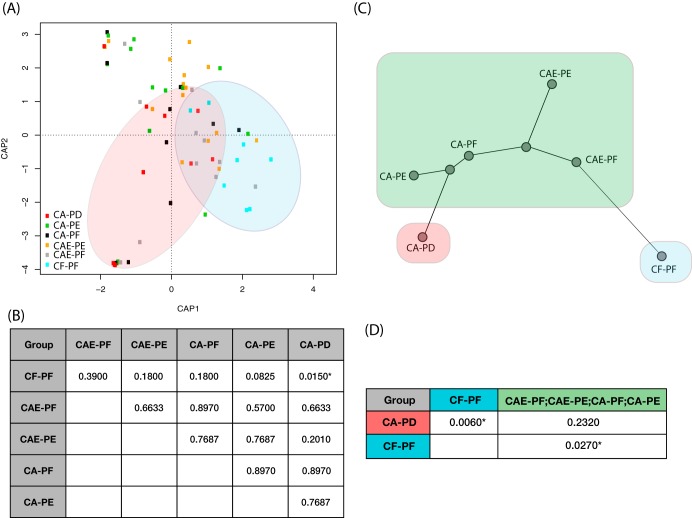
Pairwise permutational multivariate analysis of variance (PERMANOVA) tests comparing beta diversity for samples of the same plaque category at different collection time points showed no significant differences (see File S1 in the supplemental material). Consequently, we grouped all samples of the same plaque category together for all subsequent analyses. Ordination analysis showed dissimilarity between communities representing the different types of plaque (Fig. 2A). CF-PF and CAE-PF samples were mainly clustered together in the bottom right quadrant, CAE-PE samples spanned across the right and top left quadrants, and CA-PF, CA-PE, and CA-PD samples clustered mostly in the left and top left quadrants. Significant differences in beta diversity were observed between CF-PF and CA-PD samples (P = 0.015). No significant differences were observed between the remaining community comparisons (Fig. 2B). A neighbor-joining phylogeny built using pairwise PERMANOVA variance components showed increased dissimilarity in community composition as disease progressed, with CF-PF and CA-PD samples being the most dissimilar to each other (Fig. 2C). A second pairwise PERMANOVA was performed where samples categorized as CAE-PF, CAE-PE, CA-PF, and CA-PE were grouped together due to their nonsignificant differentiation in the first analysis (Fig. 2D). The community composition of CF-PF samples was significantly different from that of the grouped samples (P = 0.027), but the composition of CA-PD samples was not significantly different from the grouped samples (P = 0.232).
FIG 2.
Beta diversity analyses of oral mycobiome samples as dental caries disease progresses. (A) Constrained analysis of principal coordinates (CAP) of the different sample categories using a distance-based redundancy analysis (db-RDA). A pairwise PERMANOVA detected a significant difference in beta diversity when CF-PF samples (blue) were compared to CA-PD (red). With one exception (CA-PD), the shaded ovals show the position of CF-PF and CA-PD samples. (B) Results of pairwise PERMANOVA tests using Bray-Curtis beta diversity distances across the six different categories. Values presented are Benjamini and Hochberg FDR-corrected P values. *, statistical significance. (C) Neighbor-joining phylogeny constructed using the variance components from the pairwise PERMANOVA tests. Branch lengths are representative of the degree of difference between communities. (D) Results of pairwise PERMANOVA tests across the three significantly different tooth classification groups delineated in the initial tests (shown with red, green, and blue shading). Values presented are again Benjamini and Hochberg FDR-corrected P values. *, statistical significance.
The oral mycobiome composition in caries and dental health.
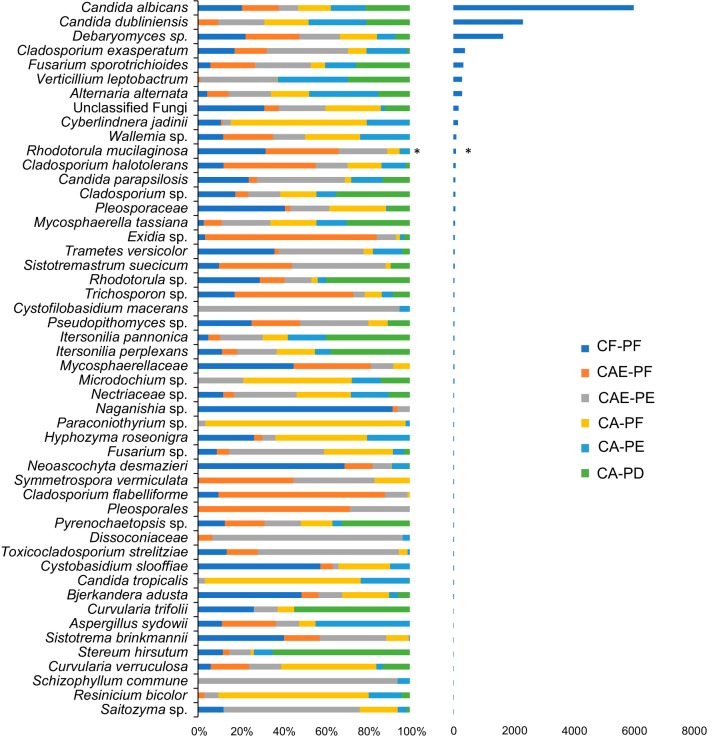
Two fungal phyla, Ascomycota and Basidiomycota, were detected in the plaque samples. All but one OTU were assigned taxonomies below the phylum level. A total of 86% of OTUs were assigned taxonomies to the genus level, and 65% of OTUs were classified to the species level. The average number of taxa per plaque sample was 21. The most abundant species across all plaque groups were C. albicans, Candida dubliniensis, Debaryomyces sp., and Cladosporium exasperatum (in order of prevalence; Fig. 3 and 4).
FIG 3.
Distribution of the top 50 most abundant taxa across the six tooth classifications using rarefied taxon counts. The left chart shows mean taxon counts for each plaque category. Means are shown as relative proportions. The chart on the right depicts the sum total of the counts for each taxon; *, taxa whose frequency was significantly different among the plaque categories (determined using a t test and adjusted for multiple comparisons using Benjamini and Hochberg FDR-corrected P values).
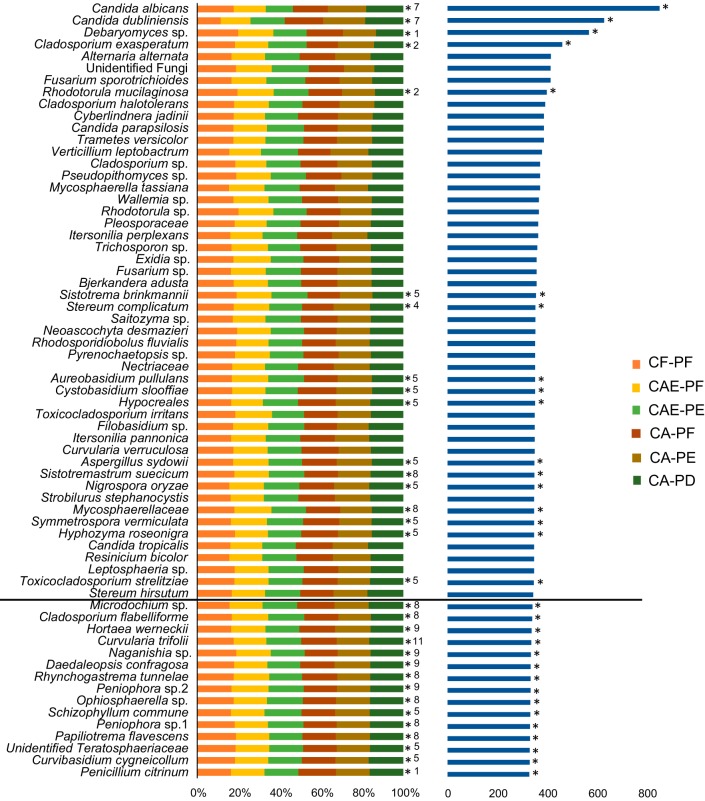
FIG 4.
Distribution of the top 50 most abundant taxa across the six plaque categories using variance-stabilizing transformation of taxon counts. The left chart shows the mean taxon counts for each plaque category. All taxa below the bold line were not present in the top 50 most abundant taxa but were statistically significantly differentially abundant. The chart on the right depicts the sum total of the counts for each taxon. *, taxa whose frequency was significantly different among the plaque categories (determined using DESeq2 and adjusted for multiple comparisons using Benjamini and Hochberg multiple testing adjustment).
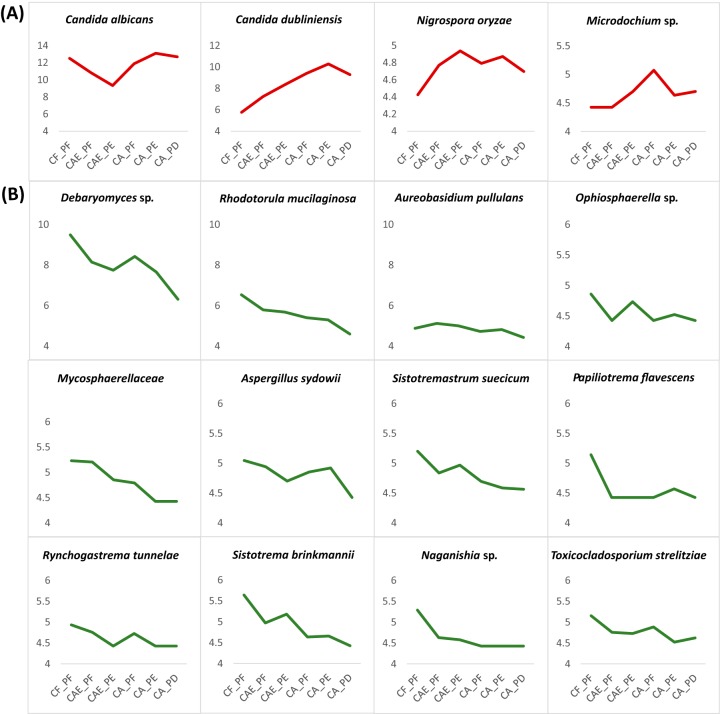
Using rarefied taxon counts, the prevalence of two taxa, Rhodotorula mucilaginosa and an unidentified Teratosphaeriaceae family, were significantly different among the plaque groups (Fig. 3) (see File S2 in the supplemental material). Using the negative binominal distribution, 32 taxa were significantly differentially abundant for at least one comparison between the plaque groups (Fig. 4). In total, there were 200 significant comparisons between the plaque groups (see File S3 in the supplemental material). The comparison between CF-PF and CA-PD showed 26 differentially abundant taxa, which was the highest number across all the comparisons. The comparison between CA-PF and CA-PE showed the lowest, with only 6 differentially abundant taxa (Table 1). Of the 32 differentially abundant taxa, C. albicans, C. dubliniensis, Nigrospora oryzae, and an unidentified Microdichium sp. were strongly correlated with caries-active plaque samples, showing statistically significant increased abundance in CA-PF, CA-PE, and CA-PD communities compared to CF-PF communities (Fig. 5A; see also Fig. S4 in the supplemental material [File S4]). C. dubliniensis showed a strong progressive increase in abundance moving from CF-PF to CA-PE, with a slight decrease moving from CA-PE to CA-PD. C. albicans showed a more variable pattern. Starting with a relatively high abundance in CF-PF, its abundance decreased until CAE-PE (this decrease from CF-PF to CAE-PE was not statistically significant [see File S4 in the supplemental material]). It then showed a similar pattern to C. dubliniensis, steadily increasing with disease progression, with a slight decrease moving from CA-PE to CA-PD. Although CF-PF abundance was relatively high, it was statistically significantly lower than both CA-PE and CA-PD. N. oryzae and Microdichium sp. did not show a steady increase with disease progression as seen for C. dubliniensis. However, both were considerably more abundant in disease than in health. N. oryzae again showed a slight decrease moving from CA-PE to CA-PD. A total of 12 differentially abundant taxa showed strong correlations with dental health (CF-PF) and showed an overall decrease in abundance as caries disease progressed. For all of these 12 taxa, their lowest abundance was observed in CA-PD (Fig. 5B).
TABLE 1.
The number of significantly differentially abundant taxa for each plaque category comparison as determined using DESeq2a
| Group | No. of significantly differentially abundant taxa for: |
||||
|---|---|---|---|---|---|
| CAE-PF | CAE-PE | CA-PF | CA-PE | CA-PD | |
| CF-PF | 11 | 9 | 13 | 16 | 26 |
| CAE-PF | 8 | 10 | 11 | 16 | |
| CAE-PE | 11 | 13 | 19 | ||
| CA-PF | 6 | 14 | |||
| CA-PE | 12 | ||||
P values were adjusted for multiple comparisons using Benjamini-Hochberg multiple testing adjustment.
FIG 5.
Significantly differentially abundant taxa enriched in dental caries and in health. (A) The four statistically significant taxa enriched in dental caries disease. (B) The 12 statistically significant taxa enriched in health. All graphs were generated from a variance-stabilizing transformation of counts within DESeq2.
The remaining 16 taxa did not show strong correlations with health or disease; however, interesting patterns were observed for these taxa. Six of these 16 taxa showed an increase in abundance in CA-PF samples, while 2 other taxa showed an increased abundance in CA-PE, subsequently followed by a decrease in abundance in CA-PD samples (see Fig. S5 in the supplemental material [File S4]). In another seven taxa, an increased abundance in CAE-PF and CAE-PE samples was observed with decreased abundance in CA-PF, CA-PE, and CA-PD samples (see Fig. S6 in the supplemental material [File S4]).
DISCUSSION
This study characterized the oral mycobiome associated with the progressive stages of caries lesions using site-specific sampling. Supragingival dental plaque samples were collected from children and tooth sites of different caries status to obtain a better understanding of how the oral mycobiome changes as disease progresses. Our findings show that fungal community composition in health-associated plaque differed significantly from that associated with dentin caries (CF-PF versus CA-PD), which is the most advanced stage of caries. Importantly, plaque from a caries-free tooth in a caries-active child (CAE-PF and CA-PF) harbored a more similar community composition to caries-associated plaque in a caries-active child (CA-PD) than to plaque from a caries-free tooth of a caries-free child (CF-PF). This suggests that healthy teeth in caries-active children are at a high risk of developing caries lesions. A previous study of ours using site-specific sampling of the bacterial community in caries showed a similar pattern (9). These findings are also concordant with studies showing that previous caries experience is an important caries risk indicator (30).
A significant difference in alpha diversity was observed only between CAE-PE and CA-PD samples; however, there was a steady decrease in community diversity as caries worsened. This pattern of decreased microbial diversity with disease has been observed previously in relation to the bacterial communities associated with dental caries and other human microbiome-associated diseases such as Crohn’s disease and ulcerative colitis (31, 32). In regard to caries, the shift toward decreased diversity is believed to be caused by increased carbohydrate consumption and fermentation leading to acid production and secretion. The low-pH environment is not optimal for the growth of many taxa, and therefore, the low pH drives the biofilm composition toward a higher abundance of acidogenic and aciduric taxa, which thrive under these conditions (32). It is possible that this is also occurring in the fungal microbiome community associated with dental caries, as some fungal taxa are known to be acidogenic and aciduric, while others are not capable of surviving in acidic environments (23, 29).
Previous studies examining the oral mycobiome in health and oral disease (caries and periodontal disease) did not observe significant differences in either alpha or beta diversity (17, 18). While their results were not statistically different, both of these studies showed a pattern of alpha diversity reduction in diseased samples. The high-resolution and site-specific nature of our sampling provides a more sensitive measure of fluctuations in community diversity, which may explain our results of statistically significant diversity compared to previous oral mycobiome studies (17, 18). It is well accepted that caries is driven by bacterial dysbiosis caused by environmental factors such as carbohydrate consumption, which leads to disruption of the plaque biofilm homeostasis toward a more pathogenic state (8, 33, 34). Our data support distinct shifts in fungal plaque communities as disease progresses, suggesting that fungal community dysbiosis is occurring and may be contributing to disease severity and progression, similar to what has been observed in bacterial dysbiosis (33–35). Furthermore, similar to bacterial communities (9), our results indicate that each stage of the disease has a characteristic taxonomic profile that is stable through time.
Only a few studies to date have utilized next-generation sequencing (NGS) of the ITS region to characterize the oral mycobiome associated with health and disease. In our study, a total of 214 OTUs comprised of 139 fungal species were identified in the dental plaque samples. This fungal diversity is consistent with previous studies that have used the ITS1 region for sequencing of the oral mycobiome (10, 17). Of note, while the number of taxa between our study and previous oral mycobiome studies that utilized the ITS1 region are similar, these other studies used oral rinse samples and not dental plaque and are therefore not directly comparable. In contrast, Fechney et al. recently characterized the oral mycobiome associated with dental caries and health in children using dental plaque samples and found a much lower community diversity (a total of 46 fungal species); however, the ITS2 region was used for sequencing and could explain these differences (18). Further comparisons need to be completed to assess variations in taxonomic resolution between the ITS1 and ITS2 regions in dental caries.
Our results support previous work highlighting the vast diversity of fungal organisms present in the oral cavity (10, 11, 17). Despite this diversity, only a relatively small number of taxa comprised the bulk of the community. Specifically, C. albicans, C. dubliniensis, Debaryomyces sp., and C. exasperatum comprised 79% of the total community, with the remainder being comprised of a large diversity of low-frequency taxa. As in other oral mycobiome studies (10, 18), C. albicans was the most abundant species in the data set and was present in 68 of the 73 samples (93.2%). C. albicans is a commensal yeast in the oral cavity and is a known opportunistic pathogen that causes diseases such as oral candidiasis (15). C. albicans has recently been gaining traction as a possible cariogenic yeast and indicator species for ECC (36–39). In many recent studies, researchers have found that the prevalence of C. albicans in children with ECC is generally much higher than in children who are caries free and that, frequently, C. albicans is detected together with and in close proximity to S. mutans (25, 27, 29, 37). Because of the high prevalence of C. albicans in the oral mucosa and in ECC, more targeted studies have been completed to determine its cariogenic potential.
Studies have shown that C. albicans is aciduric and acidogenic and is capable of biofilm formation, especially when grown in culture with S. mutans, resulting in increased amounts of extracellular polysaccharides (EPS) pertinent for caries development (23, 29, 40, 81). Recent meta-omic analyses of S. mutans and C. albicans grown in mixed biofilms have provided molecular evidence to support the hypothesis that C. albicans enhances the cariogenic potential of S. mutans (28). These results are supported by a recent study by Sampaio et al., in which they found that a dual-species biofilm composed of S. mutans and C. albicans resulted in an overall increased virulence of cariogenic biofilms compared to monospecies biofilms (41). In contrast to these studies, Fechney et al. found that there was not a significant difference in C. albicans abundance between healthy children or in children with ECC, suggesting that C. albicans is not cariogenic (18).
Our results support increased prevalence of C. albicans in CA children, possibly highlighting its role in increased caries virulence. Interestingly, our results also show a relatively high frequency of C. albicans in CF-PF children, which decreased in frequency until caries progressed into the dentin tooth layer. These results highlight the importance of site-specific sampling to gain a high-resolution profile of the mycobiome throughout disease and enable fluctuations of the mycobiome to be monitored throughout all stages of disease progression rather than just in health versus disease. To explain the pattern observed above, it is possible that multiple strains of C. albicans are present in the tooth biofilm and that as conditions begin shifting toward a more cariogenic state, one strain dies off and the other begins to proliferate. Previous studies have identified the presence of multiple genotypes of C. albicans in the oral cavity whose relative proportions can continually change after host infection to allow for biofilm adaptations to the host environment (42–44). These studies have also shown a high prevalence of C. albicans genotype A in individuals with ECC or dental caries (43, 44). To further verify that different C. albicans strains have different virulence capabilities, whole-genome sequencing and targeted biochemical tests need to be completed (45). It is important to note that, while we observed a decreased prevalence of C. albicans in the early stages of caries, it is possible that C. albicans virulence factors are still highly active and may be contributing to disease progression. C. albicans has been shown to have remarkable metabolic flexibility to utilize an array of nutrient sources and pH conditions. C. albicans is also capable of manipulating its environment to cause disease and avoid immune system detection (46). Further transcriptomic analysis is needed to fully understand the role that C. albicans contributes to caries development and progression. Our findings suggest that C. albicans may not be a reliable indicator of ECC, as it is present in high frequencies in both CF-PF children and CA-PF, CA-PE, and CA-PD children and shows a marked decrease during the initial stages of the disease.
Candida dubliniensis was the second most abundant species overall and was found in high frequency in disease. C. dubliniensis is the most closely related species to C. albicans and contains many of the same virulent traits, including the ability to produce germ tubes, chlamydospores, and hyphae (47, 48). In more recent studies, C. dubliniensis was identified in saliva samples from elderly, community-dwelling individuals and in oral wash samples from patients with chronic periodontitis, suggesting that this organism thrives when the host immune system is weakened (17). Indeed, the species is frequently found in individuals in an immunocompromised state (49).
Because of the morphological and biochemical similarities of C. albicans and C. dubliniensis, discriminating between these species in culture is problematic and, until recently, could not be done without the use of molecular technologies. It is possible that previous studies have misidentified C. dubliniensis as C. albicans and have therefore overlooked its importance in dental caries. It was only in 2015 that C. dubliniensis was first identified in plaque samples from dentinal carious lesions using scanning electron microscopy. In a study by Al-Ahmad and colleagues, C. dubliniensis and C. albicans were cultured from saliva and dental plaque samples of healthy children and children with active dentinal caries. They did not find any evidence of C. dubliniensis in CF children but found a statistically significant positive correlation between C. dubliniensis and severe childhood caries (15). Similarly, we detected low abundance/absence in CF samples, and, given that we also detected a steady increase in abundance as caries progressed, C. dubliniensis shows promise as an indicator species of ECC. However, biochemical studies and transcriptomics analyses are needed to fully elucidate the role that C. dubliniensis plays in dental caries progression and severity.
Along with C. albicans and C. dubliniensis, N. oryzae and an unidentified Microdichium sp. were also found to be associated with disease. Both of these taxa have been identified in previous studies examining the oral mycobiome of oral rinse samples. In a study by Mukherjee et al., an unidentified Nigrospora sp. was present in multiple people infected with HIV and was not observed in uninfected patients (11). In a more recent study examining the oral mycobiome in relation to periodontal disease, the taxa Nigrospora sp. and Microdichium sp. were found in higher frequency in samples from patients without periodontitis (17). Historically, based on their sexual stage, these taxa were named Khuskia sp. and Monographella sp., respectively. However, this naming system is no longer practiced (50). Future research is needed to clarify if these plant-associated pathogens are commensals in the oral cavity or if they are introduced via the environment, as well as to determine their role in dental caries.
In our study, a total of 12 taxa were associated with health and had a pattern of decreased frequency as disease progressed. Of these 12 taxa, 6 of the genera have been observed in previous oral mycobiome studies: Debaryomyces, Rhodotorula, Aureobasidium, Aspergillus, Naganishia, and Toxicocladosporium. Here, an unidentified member of the Debaryomyces genus was the third most abundant organism found overall. In previous studies, Debaryomyces hansenii has been isolated and identified from oral rinse samples, tongue scrapings, and supragingival plaque samples (13, 18). It was not statistically differentially abundant between health and disease in these studies; however, it was found in higher frequency in healthy samples. An important biochemical characteristic of D. hansenii is its ability to convert xylose (a monosaccharide in plants) to xylitol. Xylitol could potentially mitigate caries, as it negatively impacts the growth and proliferation of the cariogen S. mutans (51, 52). When xylitol is present as a carbohydrate source, S. mutans will convert it to xylitol-5-phosphate through the phosphoenolpyruvate-phosphotransferase system (PEP:PTS system). The production and accumulation of xylitol-5-phosphate within the cells inhibits glycolytic enzymes, resulting in growth inhibition, decreased metabolism, decreased acid production, and sometimes cell death (51, 52). It is possible that D. hansenii helps to maintain a healthy oral microbiome through its xylitol production and may explain its increased frequency in health (53). Further biochemical testing and metatranscriptomic analysis are needed to determine if oral fungi produce sufficient xylitol to impact cariogenic bacteria and validate a role in dental health.
R. mucilaginosa was one of the most abundant species found in our data and was significantly enriched in health. R. mucilaginosa is generally considered an opportunistic pathogen that is capable of forming biofilms and can cause a variety of infections (54). R. mucilaginosa and other unclassified members of this genus have been identified as part of the oral mycobiome in multiple other studies (11, 17, 18). In the caries mycobiome study by Fechney et al., R. mucilaginosa was not differentially abundant between health or disease; however, it was present in a higher frequency in health than in caries. R. mucilaginosa is also capable of fermenting xylose and producing xylitol, which may contribute to a role in maintaining tooth health (55).
Aureobasidium pullulans was one of the top 50 most abundant fungal taxa in the data and is another species that was strongly and significantly associated with health (showing decreased abundance as disease progressed). Members in the Aureobasidium genus, including A. pullulans, have been found to be present in the oral mycobiome in previous studies (11, 17, 18). Furthermore, this species is able to produce bioactive compounds that are useful in industry, one of which includes the production of xylanase enzymes (56). Xylanase enzymes degrade xylan to xylose, which can be fermented by other fungal members of the oral biofilm to produce xylitol. Besides producing xylanases, A. pullulans has also been found to produce liamocin oils that have strong antibacterial activity against Streptococcus species, including S. mutans (57). All of these characteristics highlight the role that A. pullulans may play in health.
Aspergillus sydowii, another species in the top 50 most abundant taxa in our data, was found to decrease in frequency in dentinal caries and was present in the highest frequency in health. Other oral mycobiome studies have identified members of the Aspergillus genus; however, none have recorded the presence of A. sydowii. Interestingly, A. sydowii is also capable of producing xylanases and many active secondary metabolites, some of which have strong antimicrobial properties, which could point to a role in dental health (58, 59).
A total of 2 remaining genera, Naganishia and Toxicocladosporium, not present in the top 50 most abundant taxa, have been isolated from the oral mycobiome in previous studies. Organisms within the genus Naganishia have been observed in the human gut, skin, and scalp as commensals, but certain members of the genus are opportunistic pathogens capable of causing disease (60). Naganishia diffluens (previously Cryptococcus diffluens) was one of the most dominant species identified by Fechney et al. in their caries mycobiome study; however, it was not associated with either health or disease (18). Similar to our results, a study completed on human fecal mycobiomes in health, ulcerative colitis, and Crohn’s disease found that Naganishia was more abundant in healthy controls than in disease (61).
The Toxicocladosporium genus was only recently described and is most often found growing on plants (62). In the Fechney et al. caries mycobiome study, they identified the presence of the fungus Toxicocladosporium rubrigenum. This species was found in higher frequencies in healthy samples (18). In our study, we identified Toxicocladosporium strelitziae in our samples, and it was also found in highest frequency in health. Future research is needed to elucidate the role that Naganishia sp. and Toxicocladosporium sp. play in caries disease; however, our results paired with previous dental caries mycobiome studies seem to implicate a role in tooth health.
The remaining taxa associated with health, Sistotrema brinkmanii, Sistotremastrum suecicum, Papiliotrema flavescens, and Rynchogastrema tunnelae, have not been identified in previous oral mycobiome studies. Most of these fungi have been isolated from the environment or from plants. More research is needed to understand if these organisms play a role in dental caries or if they are transient members of the oral mycobiota introduced via the external environment.
Finally, we observed an interesting pattern for several of the taxa associated with disease in which there was a noticeable decrease in the frequency of CA-PD. A possible explanation for this observation may be the influence of the bacterial members in the plaque biofilm. Previous research examining the bacterial microbiome associated with cavities showed that species such as Veillonella parvula, Veillonella dispar, S. mutans, Lactobacillus salivarius, and Scardovia wiggsiae increased drastically in abundance in dentinal carious lesions (9). Furthermore, both Veillonella spp. and some Lactobacillus spp. can produce acetic acid from the utilization of lactate or through heterolactic fermentation of pyruvate (63, 64). Acetic acid is known to inhibit the growth of many different types of fungi, including members of the Candida, Aspergillus, and Fusarium genera, especially when in a low-pH environment (65–67).
Another possible reason for the decreased abundance of many of the fungal species in CA-PD samples could be that they are susceptible to formic acid. Formic acid is a weak organic acid known to inhibit the growth of fungal species. Formic acid has been shown to inhibit the growth of members of the Candida, Aspergillus, and Fusarium genera at similar or greater levels than acetic acid (65, 67). A previous study examining gene expression of S. mutans when grown in culture with C. albicans showed a 5-fold increase in formate production when these two species were grown together (28). Again, it is possible that an increased abundance of S. mutans in CA-PD samples leads to increased formic acid production, creating an environment in which many fungal species cannot survive, explaining the observed pattern of decreased fungal abundance in dentinal carious lesions.
Our study utilized site-specific sampling and high-throughput amplicon sequencing to characterize the oral mycobiome through different stages of caries progression. A vast diversity of fungal organisms was identified from the supragingival plaque biofilms of children with and without ECC. We observed a strong shift in community structure for the dental mycobiome coupled with a loss of community diversity as caries progressed. C. dubliniensis, the second most abundant species in the samples, was highly correlated with disease and steadily increased in abundance as caries progressed. Our study highlights the important role it may play in disease progression and suggests that it may be a better option than C. albicans as a potential indicator species for ECC. Of note, several fungal taxa associated with health have the potential to antagonize the cariogen S. mutans via the production of xylitol and antimicrobial compounds, suggesting a possible fungal mechanism for the maintenance of dental health. Additional omic approaches, such as metatranscriptomics, and targeted biochemical studies will be required to fully explore these mechanisms and provide further insight into the complex interplay between the fungal and bacterial members of the plaque biofilm associated with caries.
MATERIALS AND METHODS
Site-specific plaque.
A total of 82 supragingival plaque samples collected from 33 children ages 2 to 7 years were included in the present study. The sampling process and the demographics of the study population are described elsewhere (68). Informed consent was obtained from the parents or legal guardians of each child, and approval for the study was granted by the Institutional Review Board of the University of Florida Health Science Center. Briefly, caries lesions were detected and diagnosed by a single examiner using the International Caries Detection and Assessment System (ICDAS-II) visual criteria (69). Lesion activity was determined by clinical appearance, plaque stagnation, and tactile sensation. Children were grouped by caries status as caries free (CF), caries active with active enamel caries lesions only (CAE), and caries active with at least two active and unrestored dentin carious lesions (CA). The range of ICDAS scores (0 to 6) as a function of caries status group were CF (for no activity, ICDAS = 0), CAE (active lesions, ICDAS, 0 to 3), and CA (active lesions, ICDAS = 0 to 6). The threshold for the CA group was the presence of at least two ICDAS scores of 5 or 6 (cavitated dentin lesions). Supragingival plaque samples were collected from separate tooth surfaces and classified as follows: PF for samples collected from tooth surfaces with no active carious lesions (ICDAS score = 0), PE for samples collected from an enamel carious lesion (ICDAS score = 1 to 3), or PD for samples collected from dentin carious lesions (ICDAS score = 4 to 6). Multiple plaque samples were collected from children who had teeth with more than one plaque category. Combining caries status with tooth classification produces plaque categories representing six progressive stages of the disease: CF-PF (n = 8), CAE-PF (n = 12), CAE-PE (n = 16), CA-PF (n = 13), CA-PE (n = 16), and CA-PD (n = 17) (see File S5 in the supplemental material).
DNA extraction and library construction.
Fungal genomic DNA was extracted using the Qiagen DNeasy PowerBiofilm kit (Qiagen, USA) according to the manufacturer’s protocol. An extraction blank was included in each set of extractions to monitor for external contamination.
The fungal ITS1 region was amplified using PCR. The forward primer used was ITS1F (CTTGGTCATTTAGAGGAAGTAA), and the reverse primer used was ITS2 (GCTGCGTTCTTCATCGATGC) (10). Custom-barcoded primers were designed using the fungal ITS1 primers as outlined previously in Kozich et al. (70). Positive and negative control samples were run with each PCR. After PCR amplification, samples were run on 2.0% agarose Tris-acetate-EDTA (TAE) gels. The amplicon band was gel extracted and purified using Qiagen’s QIAquick gel extraction kit (Qiagen, USA) according to the manufacturer’s protocol. The concentration of the gel-extracted samples was measured using Qubit 3.0 (Thermo Fisher, USA). Samples were prepared for sequencing following Illumina’s “Denature and Dilute Libraries Guide” (https://support.illumina.com/content/dam/illumina-support/documents/documentation/system_documentation/miseq/miseq-denature-dilute-libraries-guide-15039740-10.pdf) and the Illumina “16S Metagenomic Sequencing Library Preparation Guide” (https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf). Custom read 1, read 2, and index primers were spiked into the Illumina reagent cartridge as outlined previously (71).
Illumina sequencing, sequence analysis, and taxonomic classification.
Fungal amplicons were sequenced on Illumina’s MiSeq platform (Illumina, San Diego, CA) using a V2 500-cycle kit (250-bp paired-end reads). Demultiplexed paired-end reads were imported into QIIME2 (72) for sequence analysis. Sequence quality control was completed using the DADA2 pipeline incorporated into QIIME2. This program filtered and trimmed reads, removed chimeras, assigned reads to OTUs, and generated an OTU table. To obtain taxonomic assignment of the reads post-quality control, the dynamic UNITE fungal database with a 97 to 99% similarity threshold was used to fit and train a naive-Bayes taxonomic classifier. After taxonomic classification, multiple OTU IDs were often assigned to the same taxon. Using QIIME2, we collapsed OTUs with the same taxonomic classification at the species level.
Statistical analysis.
Prior to completing diversity estimates, samples were rarefied to an even sampling depth. The samples did not undergo any further preprocessing to remove singletons or low-abundance OTUs before completing alpha diversity analysis. Prior to completing beta diversity analysis, sequences were filtered to remove singletons and OTUs present in less than 0.1% total abundance.
Statistical analysis of the sequences was completed using the QIIME2 pipeline (version 2019.1) (73), QIIME1 (version 1.9.0) (74), and R Studio (version 1.1.463) (75). The following packages were used in R Studio: phyloseq (version 1.26.1) (76), qiime2R (version 0.99.11), DESeq2 (version 1.22.2) (77), and microbiomeSeq (version 0.1) (73). The rarefied count table was used to estimate alpha and beta diversity metrics. Sampling depth was set to 2,194 reads, as this was the sample that had the lowest number of taxonomically assigned reads. Alpha diversity was estimated in QIIME2 using the Shannon index (78). Statistical significance of alpha diversity was calculated using nonparametric, pairwise Kruskal-Wallis tests. Adjusted P values were calculated using Benjamini and Hochberg false-discovery rate (FDR) correction for multiple comparisons (79). Alpha diversity plots were generated using the phyloseq package in R. Beta diversity was estimated in QIIME2 using the Bray-Curtis index (80). Pairwise PERMANOVAs were completed to determine statistical differences in beta diversity. Adjusted P values were calculated using Benjamini and Hochberg correction for multiple comparisons. Results were imported into QIIME1 to generate constrained analysis of principal coordinate (CAP) plots using distance-based redundancy analysis (db-RDA).
Differential abundance testing was completed using both QIIME1 and DESeq2 in phyloseq. In the QIIME1 analysis, Kruskal-Wallis tests were completed on rarefied data using the compare_categories.py script to determine significant differences in OTU frequency between different plaque categories. P values were generated using 10,000 permutations and were corrected for using the FDR. McMurdie and Holmes addressed the loss of statistical power when rarefying data for use in differential abundance testing and proposed a more sensitive method for detecting differentially abundant taxa through the use of the negative binomial distribution as implemented in DESeq2 (77). Here, we also take this approach to determine differentially abundant OTUs among different plaque sample categorizations. The nonrarefied and unfiltered read table generated in QIIME2 after DADA2 quality control was used for this analysis. The data were log transformed using variance-stabilizing transformation (VSD) to generate OTU frequencies. P values were corrected for using Benjamini and Hochberg FDR correction for multiple comparisons.
To generate taxonomic profiles for the most abundant taxa, both the rarified OTU table from QIIME and the VSD normalized table from DESeq2 were used. The top 50 most abundant taxa were plotted using Excel for each analysis. The overall relative abundance of the top 50 taxa across the data set was also plotted.
Data availability.
Sequence data have been deposited in the NCBI database under the BioProject accession number PRJNA589851.
Ethics approval and consent to participate.
Informed consent was obtained from the parents or legal guardians for each child in the study, and the protocol was approved by the Institutional Review Board of the University of Florida Health Science Center (UF IRB, 201600154).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Clemson University Biological Sciences Department, specifically the Clemson University’s biological sciences graduate professional development course, Grants in Aid of Research Grant, for their funding support for this work.
L.M.O. and V.P.R. conceptualized this project and its experimental design. M.M.N. developed and completed sample collection and sample collection methods. L.M.O., R.S., and G.S. completed sample processing. L.M.O. completed sequencing and data analyses. L.M.O., V.P.R., M.M.N., and R.A.B. contributed to manuscript revisions and improvements of the final manuscript. All authors have read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Phantumvanit P, Makino Y, Ogawa H, Rugg-Gunn A, Moynihan P, Petersen PE, Evans W, Feldens CA, Lo E, Khoshnevisan MH, Baez R, Varenne B, Vichayanrat T, Songpaisan Y, Woodward M, Nakornchai S, Ungchusak C. 2018. WHO Global Consultation on Public Health Intervention against Early Childhood Caries. Community Dent Oral Epidemiol 46:280–287. doi: 10.1111/cdoe.12362. [DOI] [PubMed] [Google Scholar]
- 2.Twetman S. 2018. Prevention of dental caries as a non-communicable disease. Eur J Oral Sci 1:19–25. doi: 10.1111/eos.12528. [DOI] [PubMed] [Google Scholar]
- 3.Anil S, Anand PS. 2017. Early childhood caries: prevalence, risk factors, and prevention. Front Pediatr 5:157. doi: 10.3389/fped.2017.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, Romero H, Simón-Soro A, Pignatelli M, Mira A. 2012. The oral metagenome in health and disease. ISME J 6:46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jagathrakshakan SN, Sethumadhava RJ, Mehta DT, Ramanathan A. 2015. 16S rRNA gene-based metagenomic analysis identifies a novel bacterial co-prevalence pattern in dental caries. Eur J Dent 9:127–132. doi: 10.4103/1305-7456.149661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loesche WJ. 1986. Role of Streptococcus mutans in human dental decay. Microbiol Rev 50:353–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, Leys EJ, Paster BJ. 2008. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol 46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi N, Nyvad B. 2011. The role of bacteria in the caries process: ecological perspectives. J Dent Res 90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- 9.Richards VP, Alvarez AJ, Luce AR, Bedenbaugh M, Mitchell ML, Burne RA, Nascimento MM. 2017. Microbiomes of site-specific dental plaques from children with different caries status. Infect Immun 85:e00106-17. doi: 10.1128/IAI.00106-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. 2010. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukherjee PK, Chandra J, Retuerto M, Sikaroodi M, Brown RE, Jurevic R, Salata RA, Lederman MM, Gillevet PM, Ghannoum MA. 2014. Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog 10:e1003996. doi: 10.1371/journal.ppat.1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA, Dongari-Bagtzoglou A, Diaz PI, Strausbaugh LD. 2014. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One 9:e90899. doi: 10.1371/journal.pone.0090899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ieda S, Moriyama M, Takeshita T, Takashita T, Maehara T, Imabayashi Y, Shinozaki S, Tanaka A, Hayashida J-N, Furukawa S, Ohta M, Yamashita Y, Nakamura S. 2014. Molecular analysis of fungal populations in patients with oral candidiasis using internal transcribed spacer region. PLoS One 9:e101156. doi: 10.1371/journal.pone.0101156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kneist S, Borutta A, Sigusch BW, Nietzsche S, Küpper H, Kostrzewa M, Callaway A. 2015. First-time isolation of Candida dubliniensis from plaque and carious dentine of primary teeth. Eur Arch Paediatr Dent 16:365–370. doi: 10.1007/s40368-015-0180-1. [DOI] [PubMed] [Google Scholar]
- 15.Al-Ahmad A, Auschill TM, Dakhel R, Wittmer A, Pelz K, Heumann C, Hellwig E, Arweiler NB. 2016. Prevalence of Candida albicans and Candida dubliniensis in caries-free and caries-active children in relation to the oral microbiota—a clinical study. Clin Oral Invest 20:1963–1971. doi: 10.1007/s00784-015-1696-9. [DOI] [PubMed] [Google Scholar]
- 16.Zakaria MN, Furuta M, Takeshita T, Shibata Y, Sundari R, Eshima N, Ninomiya T, Yamashita Y. 2017. Oral mycobiome in community-dwelling elderly and its relation to oral and general health conditions. Oral Dis 23:973–982. doi: 10.1111/odi.12682. [DOI] [PubMed] [Google Scholar]
- 17.Peters BA, Wu J, Hayes RB, Ahn J. 2017. The oral fungal mycobiome: characteristics and relation to periodontitis in a pilot study. BMC Microbiol 17:157. doi: 10.1186/s12866-017-1064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fechney JM, Browne GV, Prabhu N, Irinyi L, Meyer W, Hughes T, Bockmann M, Townsend G, Salehi H, Adler CJ. 2019. Preliminary study of the oral mycobiome of children with and without dental caries. J Oral Microbiol 11:1536182. doi: 10.1080/20002297.2018.1536182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoch CL, Fungal Barcoding Consortium Author List, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Bolchacova E, Voigt K, Crous PW, Miller AN, Wingfield MJ, Aime MC, An K-D, Bai F-Y, Barreto RW, Begerow D, Bergeron M-J, Blackwell M, Boekhout T, Bogale M, Boonyuen N, Burgaz AR, Buyck B, Cai L, Cai Q, Cardinali G, Chaverri P, Coppins BJ, Crespo A, Cubas P, Cummings C, Damm U, de Beer ZW, de Hoog GS, Del-Prado R, Dentinger B, Dieguez-Uribeondo J, Divakar PK, Douglas B, Duenas M, Duong TA, Eberhardt U, Edwards JE, Elshahed MS, Fliegerova K, Furtado M, Garcia MA, Ge Z-W. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci U S A 109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas A, Mhambrey S, Chokshi K, Chokshi A, Jana S, Thakur S, Jose D, Bajpai G. 2016. Association of oral Candida albicans with severe early childhood caries – a pilot study. J Clin Diagn Res 10:ZC109–ZC112. doi: 10.7860/JCDR/2016/19387.8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peretz B, Mazor Y, Dagon N, Bar-Ness Greenstein R. 2011. Candida, mutans streptococci, oral hygiene and caries in children. J Clin Pediatr Dent 36:185–188. doi: 10.17796/jcpd.36.2.f1m4283501374t22. [DOI] [PubMed] [Google Scholar]
- 22.Neves A, Lobo L, Pinto K, Pires E, Requejo M, Maia L, Antonio A. 2015. Comparison between clinical aspects and salivary microbial profile of children with and without early childhood caries: a preliminary study. J Clin Pediatr Dent 39:209–214. doi: 10.17796/1053-4628-39.3.209. [DOI] [PubMed] [Google Scholar]
- 23.Klinke T, Guggenheim B, Klimm W, Thurnheer T. 2011. Dental caries in rats associated with Candida albicans. Caries Res 45:100–106. doi: 10.1159/000324809. [DOI] [PubMed] [Google Scholar]
- 24.de Carvalho FG, Silva DS, Hebling J, Spolidorio LC, Spolidorio D. 2006. Presence of mutans streptococci and Candida spp. in dental plaque/dentine of carious teeth and early childhood caries. Arch Oral Biol 51:1024–1028. doi: 10.1016/j.archoralbio.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Raja M, Hannan A, Ali K. 2010. Association of oral candidal carriage with dental caries in children. Caries Res 44:272–276. doi: 10.1159/000314675. [DOI] [PubMed] [Google Scholar]
- 26.Xiao J, Grier A, Faustoferri RC, Alzoubi S, Gill AL, Feng C, Liu Y, Quivey RG, Kopycka-Kedzierawski DT, Koo H, Gill SR. 2018. Association between oral Candida and bacteriome in children with severe ECC. J Dent Res 97:1468–1476. doi: 10.1177/0022034518790941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai C-H, Gonzalez-Begne M, Watson G, Krysan DJ, Bowen WH, Koo H. 2014. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun 82:1968–1981. doi: 10.1128/IAI.00087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He J, Kim D, Zhou X, Ahn SJ, Burne RA, Richards VP, Koo H. 2017. RNA-seq reveals enhanced sugar metabolism in Streptococcus mutans co-cultured with Candida albicans within mixed-species biofilms. Front Microbiol 8:1036. doi: 10.3389/fmicb.2017.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao J, Huang X, Alkhers N, Alzamil H, Alzoubi S, Wu TT, Castillo DA, Campbell F, Davis J, Herzog K, Billings R, Kopycka-Kedzierawski DT, Hajishengallis E, Koo H. 2018. Candida albicans and early childhood caries: a systematic review and meta-analysis. Caries Res 52:102–112. doi: 10.1159/000481833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee HJ, Kim JB, Jin BH, Paik D Il, Bae KH. 2015. Risk factors for dental caries in childhood: a five-year survival analysis. Community Dent Oral Epidemiol 43:163–171. doi: 10.1111/cdoe.12136. [DOI] [PubMed] [Google Scholar]
- 31.Dalal SR, Chang EB. 2014. The microbial basis of inflammatory bowel diseases. J Clin Invest 124:4190–4196. doi: 10.1172/JCI72330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valm AM. 2019. The structure of dental plaque microbial communities in the transition from health to dental caries and periodontal disease. J Mol Biol 431:2957–2969. doi: 10.1016/j.jmb.2019.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamont RJ, Koo H, Hajishengallis G. 2018. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol 16:745–759. doi: 10.1038/s41579-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marsh PD, Zaura E. 2017. Dental biofilm: ecological interactions in health and disease. J Clin Periodontol 44:S12–S22. doi: 10.1111/jcpe.12679. [DOI] [PubMed] [Google Scholar]
- 35.Bowen WH, Burne RA, Wu H, Koo H. 2018. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol 26:229–242. doi: 10.1016/j.tim.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De-la-Torre J, Marichalar-Mendia X, Varona-Barquin A, Marcos-Arias C, Eraso E, Aguirre-Urizar JM, Quindós G. 2016. Caries and Candida colonisation in adult patients in Basque country (Spain). Mycoses 59:234–240. doi: 10.1111/myc.12453. [DOI] [PubMed] [Google Scholar]
- 37.Xiao J, Moon Y, Li L, Rustchenko E, Wakabayashi H, Zhao X, Feng C, Gill SR, McLaren S, Malmstrom H, Ren Y, Quivey R, Koo H, Kopycka-Kedzierawski DT. 2016. Candida albicans carriage in children with severe early childhood caries (S-ECC) and maternal relatedness. PLoS One 11:e0164242. doi: 10.1371/journal.pone.0164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lozano Moraga CP, Rodríguez Martínez GA, Lefimil Puente CA, Morales Bozo IC, Urzúa Orellana BR. 2017. Prevalence of Candida albicans and carriage of Candida non-albicans in the saliva of preschool children, according to their caries status. Acta Odontol Scand 75:30–35. doi: 10.1080/00016357.2016.1244560. [DOI] [PubMed] [Google Scholar]
- 39.Pereira DFA, Seneviratne CJ, Koga-Ito CY, Samaranayake LP. 2018. Is the oral fungal pathogen Candida albicans a cariogen? Oral Dis 24:518–526. doi: 10.1111/odi.12691. [DOI] [PubMed] [Google Scholar]
- 40.Klinke T, Kneist S, De Soet JJ, Kuhlisch E, Mauersberger S, Förster A, Klimm W. 2009. Acid production by oral strains of Candida albicans and Lactobacilli. Caries Res 43:83–91. doi: 10.1159/000204911. [DOI] [PubMed] [Google Scholar]
- 41.Sampaio AA, Souza SE, Ricomini-Filho AP, Del Bel Cury AA, Cavalcanti YW, Cury JA. 2019. Candida albicans increases dentine demineralization provoked by Streptococcus mutans biofilm. Caries Res 53:322–331. doi: 10.1159/000494033. [DOI] [PubMed] [Google Scholar]
- 42.Sampaio P, Gusmão L, Correia A, Alves C, Rodrigues AG, Pina-Vaz C, Amorim A, Pais C. 2005. New microsatellite multiplex PCR for Candida albicans strain typing reveals microevolutionary changes. J Clin Microbiol 43:3869–3876. doi: 10.1128/JCM.43.8.3869-3876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu N, Lin J, Wu L, Zhao J. 2015. Distribution of Candida albicans in the oral cavity of children aged 3–5 years of Uygur and Han nationality and their genotype in caries-active groups. Genet Mol Res 14:748–757. doi: 10.4238/2015.January.30.18. [DOI] [PubMed] [Google Scholar]
- 44.da Silva-Rocha WP, Lemos V, Svidizisnki TIE, Milan EP, Chaves GM. 2014. Candida species distribution, genotyping and virulence factors of Candida albicans isolated from the oral cavity of kidney transplant recipients of two geographic regions of Brazil. BMC Oral Health 14:20. doi: 10.1186/1472-6831-14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W, Li Y, Lin J, Abduryim A, Zhao J. 2018. Cariogenicity of Candida albicans of distinct genotypes among 3-5-year-old Uygur children in Kashgar, China- a case-control study. BMC Oral Health 18. doi: 10.1186/s12903-018-0658-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miramon P, Lorenz MC. 2017. A feast for Candida: metabolic plasticity confers an edge for virulence. PLoS Pathog 13:e1006144. doi: 10.1371/journal.ppat.1006144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson AP, Gamble JA, Yeomans T, Moran GP, Saunders D, Harris D, Aslett M, Barrell JF, Butler G, Citiulo F, Coleman DC, De Groot PWJ, Goodwin TJ, Quail MA, McQuillan J, Munro CA, Pain A, Poulter RT, Rajandream MA, Renauld H, Spiering MJ, Tivey A, Gow NAR, Barrell B, Sullivan DJ, Berriman M. 2009. Comparative genomics of the fungal pathogens Candida dubliniensis and Candida albicans. Genome Res 19:2231–2244. doi: 10.1101/gr.097501.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar K, Prakash A, Tasleem M, Islam A, Ahmad F, Hassan MI. 2014. Functional annotation of putative hypothetical proteins from Candida dubliniensis. Gene 543:93–100. doi: 10.1016/j.gene.2014.03.060. [DOI] [PubMed] [Google Scholar]
- 49.Chang EY, Fatima S, Balan S, Bhyravabhotla K, Erickson M, Chan A, Ivonye C, Bradley C. 2018. Candida dubliniensis abscess: a clinical case and a review of the literature. Med Mycol Case Rep 21:41–43. doi: 10.1016/j.mmcr.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raja HA, Miller AN, Pearce CJ, Oberlies NH. 2017. Fungal identification using molecular tools: a primer for the natural products research community. J Nat Prod 80:756–770. doi: 10.1021/acs.jnatprod.6b01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Söderling EM. 2009. Xylitol, mutans streptococci, and dental plaque. Adv Dent Res 21:74–78. doi: 10.1177/0895937409335642. [DOI] [PubMed] [Google Scholar]
- 52.Janakiram C, Deepan Kumar CV, Joseph J. 2017. Xylitol in preventing dental caries: a systematic review and meta-analyses. J Nat Sci Biol Med 8:16–21. doi: 10.4103/0976-9668.198344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.López-Linares JC, Romero I, Cara C, Castro E, Mussatto SI. 2018. Xylitol production by Debaryomyces hansenii and Candida guilliermondii from rapeseed straw hemicellulosic hydrolysate. Bioresour Technol 247:736–743. doi: 10.1016/j.biortech.2017.09.139. [DOI] [PubMed] [Google Scholar]
- 54.Wirth F, Goldani LZ. 2012. Epidemiology of rhodotorula: an emerging pathogen. Interdiscip Perspect Infect Dis 2012:465717. doi: 10.1155/2012/465717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bura R, Vajzovic A, Doty SL. 2012. Novel endophytic yeast Rhodotorula mucilaginosa strain PTD3 I: production of xylitol and ethanol. J Ind Microbiol Biotechnol 39:1003–1011. doi: 10.1007/s10295-012-1109-x. [DOI] [PubMed] [Google Scholar]
- 56.Chi Z, Wang F, Chi Z, Yue L, Liu G, Zhang T. 2009. Bioproducts from Aureobasidium pullulans, a biotechnologically important yeast. Appl Microbiol Biotechnol 82:793–804. doi: 10.1007/s00253-009-1882-2. [DOI] [PubMed] [Google Scholar]
- 57.Bischoff KM, Leathers TD, Price NP, Manitchotpisit P. 2015. Liamocin oil from Aureobasidium pullulans has antibacterial activity with specificity for species of Streptococcus. J Antibiot (Tokyo) 68:642–645. doi: 10.1038/ja.2015.39. [DOI] [PubMed] [Google Scholar]
- 58.Nair SG, Sindhu R, Shashidhar S. 2008. Purification and biochemical characterization of two xylanases from Aspergillus sydowii SBS 45. Appl Biochem Biotechnol 149:229–243. doi: 10.1007/s12010-007-8108-9. [DOI] [PubMed] [Google Scholar]
- 59.Wang JF, Lin XP, Qin C, Liao SR, Wan JT, Zhang TY, Liu J, Fredimoses M, Chen H, Yang B, Zhou XF, Yang XW, Tu ZC, Liu YH. 2014. Antimicrobial and antiviral sesquiterpenoids from sponge-associated fungus, Aspergillus sydowii ZSDS1-F6. J Antibiot (Tokyo) 67:581–583. doi: 10.1038/ja.2014.39. [DOI] [PubMed] [Google Scholar]
- 60.Inácio J, Daniel HM. 2017. Commensalism: the case of the human zymobiome. Ecology 2017:211–228. doi: 10.1007/978-3-319-61575-2_8. [DOI] [Google Scholar]
- 61.Imai T, Inoue R, Kawada Y, Morita Y, Inatomi O, Nishida A, Bamba S, Kawahara M, Andoh A. 2019. Characterization of fungal dysbiosis in Japanese patients with inflammatory bowel disease. J Gastroenterol 54:149–159. doi: 10.1007/s00535-018-1530-7. [DOI] [PubMed] [Google Scholar]
- 62.Bezerra JDP, Sandoval-Denis M, Paiva LM, Silva GA, Groenewald JZ, Souza-Motta CM, Crous PW. 2017. New endophytic Toxicocladosporium species from cacti in Brazil, and description of Neocladosporium gen. nov. IMA Fungus 8:77–97. doi: 10.5598/imafungus.2017.08.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Badet C, Thebaud NB. 2008. Ecology of lactobacilli in the oral cavity: a review of literature. Open Microbiol J 2:38–48. doi: 10.2174/1874285800802010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu S, Chen M, Wang Y, Zhou X, Peng X, Ren B, Li M, Cheng L. 2020. Effect of Veillonella parvula on the physiological activity of Streptococcus mutans. Arch Oral Biol 109:104578. doi: 10.1016/j.archoralbio.2019.104578. [DOI] [PubMed] [Google Scholar]
- 65.Hassan RA, Sand MI, El-Kadi SHM. 2012. Effect of some organic acids on fungal growth and their toxins production. J Agric Chem Biotechnol 3:391–397. doi: 10.21608/JACB.2012.55011. [DOI] [Google Scholar]
- 66.Lourenço A, Pedro NA, Salazar SB, Mira NP. 2018. Effect of acetic acid and lactic acid at low pH in growth and azole resistance of Candida albicans and Candida glabrata. Front Microbiol 9:3265. doi: 10.3389/fmicb.2018.03265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lastauskienė E, Zinkevičienė A, Girkontaitė I, Kaunietis A, Kvedarienė V. 2014. Formic acid and acetic acid induce a programmed cell death in pathogenic Candida species. Curr Microbiol 69:303–310. doi: 10.1007/s00284-014-0585-9. [DOI] [PubMed] [Google Scholar]
- 68.Nascimento MM, Alvarez AJ, Huang X, Hanway S, Perry S, Luce A, Richards VP, Burne RA. 2019. Arginine metabolism in supragingival oral biofilms as a potential predictor of caries risk. JDR Clin Trans Res 4:262–270. doi: 10.1177/2380084419834234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ekstrand KR, Martignon S, Ricketts DJN, Qvist V. 2007. Detection and activity assessment of primary coronal caries lesions: a methodologic study. Oper Dent 32:225–235. doi: 10.2341/06-63. [DOI] [PubMed] [Google Scholar]
- 70.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rideout JR, Dillon MR, Bokulich NA, Abnet C, Ghalith GA, Alexander H, Alm EJ, Arumugam M, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Colin J, Brown CT, Callahan BJ, Mauricio A, Rodríguez C, Chase J, Cope E, Da Silva R, Dorrestein PC, Douglas GM, Duvallet C, Edwardson CF, Ernst M, Fouquier J, Gauglitz JM, Gibson DL, Gonzalez A, Huttley GA, Janssen S, Jarmusch AK, Kaehler BD, Kang K, Bin Keefe CR, Keim P, Kelley ST, Ley R, Loftfield E, Marotz C, Martin B, Mcdonald D, Mciver LJ, Alexey V, Metcalf JL, Morgan SC, Morton JT, Naimey AT. 2018. QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Prepr 6:e27295v2. doi: 10.7287/peerj.preprints.27295v2. [DOI] [Google Scholar]
- 73.Ssekagiri A, Sloan WT, Ijaz UZ. 2017. microbiomeSeq: an R package for analysis of microbial communities in an environmental context. In ISCB Africa ASBCB Conference, Kumasi, Ghana: https://github.com/umerijaz/microbiomeSeq. [Google Scholar]
- 74.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.RStudio Team. 2015. RStudio: integrated development for R. RStudio, Inc., Boston, MA: https://rstudio.com/. [Google Scholar]
- 76.McMurdie PJ, Holmes S. 2013. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shannon CE. 1948. A mathematical theory of communication. Bell Syst Tech J 27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 79.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 80.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 81.Ellepola K, Truong T, Liu Y, Lin Q, Lim TK, Lee YM, Cao T, Koo H, Seneviratne CJ. 2019. Multi-omics analyses reveal synergistic carbohydrate metabolism in Streptococcus mutans-Candida albicans mixed-species biofilms. Infect Immun 87:e00339-19. doi: 10.1128/IAI.00339-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data have been deposited in the NCBI database under the BioProject accession number PRJNA589851.