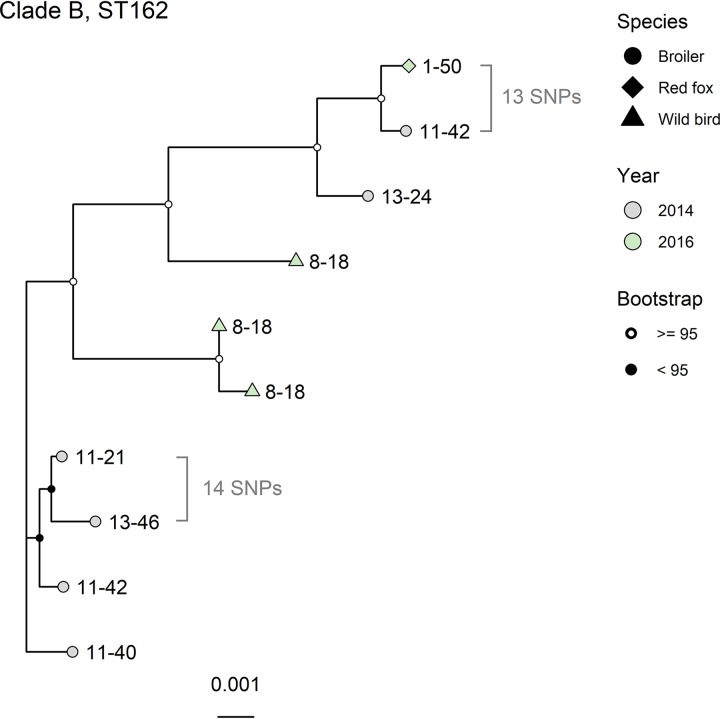
FIG 3.
Maximum likelihood core-genome tree of clade B, containing 10 ST162 isolates. Tip labels denote the location of the isolate by county-municipality. Core-genome SNPs were called with ParSNP, recombinant sites were removed with Gubbins, and the tree was generated with IQTree. The evolutionary model used was TIMe+ASC+R2. The percentage of the genome shared among all isolates was 86%. The highly similar isolates from wild birds in this tree (location 8-18, 2016) were disregarded as they were from the same sample, one isolated by the traditional method and the other isolated by the selective method.