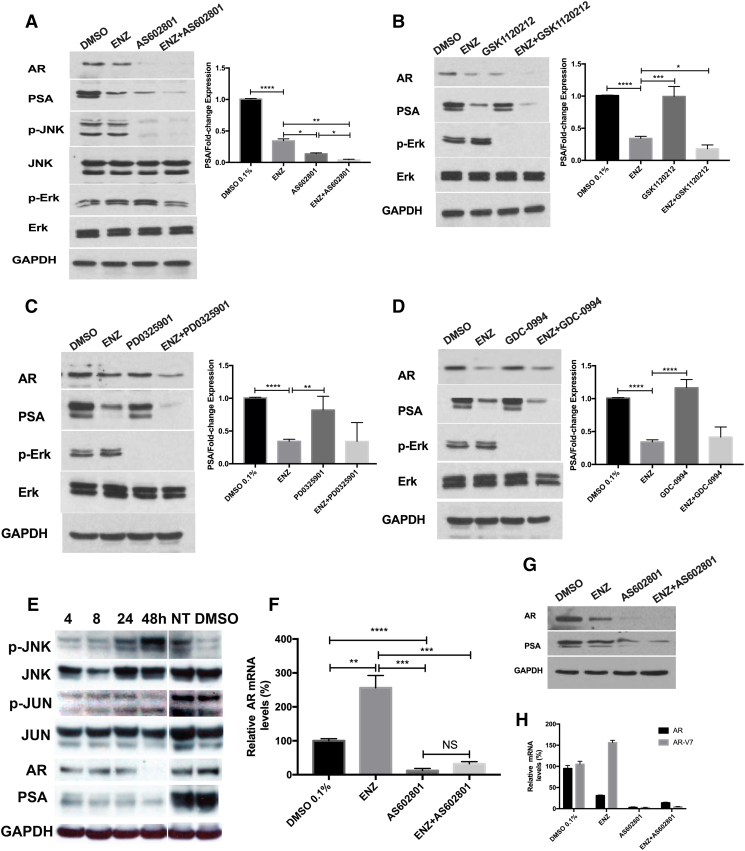
Figure 4.
Effect of Enzalutamide and/or AS602801, GSK1120212, PD-0325901, GDC-0994 on pathway protein and mRNA expression. (A-D) LNCaP cells (1×106 cells) were seeded in 6-well plates and incubated for 24 h to allow for adherence. Cells were treated with DMSO 0.1% (control); enzalutamide (12.5 μM); AS602801 (7.5 μM); GSK1120212 (12.5 μM); PD-0325901(25 μM); GDC-0994 (1 μM) alone and combination 48 h for PSA and AR detection. For p-Erk detection, LNCaP cells were pretreated with 5 μg/ml EGF for 1 h after 24 h starvation, then treated with indicated concentration for 1 h. For p-JNK detection, LNCaP cells were treated with indicated concentration for 2 hours, then stimulated with 25 μg/ml Anisomycin for 30mins. Western blot analysis was conducted for AR, PSA, p-ERK(Thr202/Tyr204)/ERK, p-JNK/JNK. GAPDH was used as loading control. (E) LNCaP cells were treated with AS602801 (7.5 μM) for various time points. AR/PSA, JNK, p-JNK, and p-c-Jun/c-Jun were analyzed by Western blot analysis; GAPDH was using as a loading control. NT indicates untreated LNCaP cells, and DMSO indicates LNCaP cells treated with vehicle for 48 hours. The expression level of PSA was quantified with Fiji. (F) LNCaP cells were treated with indicated concentrations for 48 h for RNA extraction and analyzed by QPCR. AR mRNA levels were normalized by co-amplification of beta-actin mRNA and are shown as relative fold change. (G) C4-2 cells were treated with DMSO 0.1% and enzalutamide (12.5 μM); AS602801 (7.5 μM) and combination. AR and PSA were analyzed by Western blot analysis; GAPDH was using as a loading control. (H) C4-2 cells were treated with indicated concentrations for 48 h for RNA extraction and analyzed by QPCR. AR and AR-V7 mRNA levels were normalized by co-amplification of beta-actin mRNA and are shown as relative fold change. All the experiments were repeated three times, and the data are presented as the mean ± SD. *P < .1, **P < .01, ***P < .001, ****P < .0001.