Short abstract
Prohibitin 1 is an evolutionary conserved and ubiquitously expressed protein that exerts different biological functions depending on its subcellular localization. The role of prohibitin 1 in liver cancer is controversial as it can be pro- or anti-tumorigenic. However, most of the studies to date have described prohibitin 1 primarily as a tumor suppressor in the liver. Its deficiency sensitizes the liver to cholestatic liver injury, non-alcoholic fatty liver disease, inflammatory insults, and cancer. Liver-specific Phb1-knockout mice spontaneously develop hepatocellular carcinoma, Phb1 heterozygotes are more susceptible to develop cholangiocarcinoma, and the majority of human hepatocellular carcinomas and cholangiocarcinomas have reduced prohibitin 1 expression. Consistent with a tumor suppressive role in the liver, prohibitin 1 negatively regulates proliferation in hepatocytes and human hepatocellular carcinoma and cholangiocarcinoma cell lines, and multiple oncogenic signaling pathways are activated when prohibitin 1 is deficient. Although best known as a mitochondrial chaperone, prohibitin 1 can protect the liver by mitochondrial-independent mechanisms. This review summarizes what’s known about prohibitin 1’s role in liver pathology, with the focus on hepatoprotection and carcinogenesis.
Impact statement
This review summarizes the last decades of research on PHB1 in liver pathobiology. PHB1 is a key player for liver health as it is hepatoprotective and tumor suppressive. We highlight the importance of PHB1’s subcellular localization, post-translational modifications, and interacting proteins as major determinants of PHB1 cytoprotective function and anti-tumor activity in the liver.
Keywords: Prohibitin, hepatocellular carcinoma, cholangiocarcinoma, cholestatic liver injury
Introduction
Prohibitin 1 (PHB1) is an evolutionary conserved and ubiquitously expressed protein with multiple biological functions related to its subcellular localizations, post-translational modifications (PTMs), and interactome.1 PHB1 is found in the mitochondria, nucleus, and plasma membrane and regulates mitochondrial function, gene expression, and cell signaling, respectively.2 PHB1 was originally isolated from regenerating rat liver and characterized as an anti-proliferative gene (hence its name).3 However, its role in cancer is controversial since both anti-tumorigenic and pro-tumorigenic effects have been reported even in the same organ.1 In the liver, PHB1 acts mainly as a tumor suppressor and its deficiency results in liver injury and cancer.4–9 This review examines the role of PHB1 in liver pathology focusing on the how its subcellular localizations, PTMs, and interactome may regulate its different functions.
Different functions and subcellular localizations of PHB1
Several studies have attributed the multifunctionality of PHB1 to its diverse subcellular localizations.1,2,10 PHB1’s specific structure allows it to shuttle between subcellular compartments. It has an N-terminal hydrophobic alpha helix domain that acts as a membrane-anchor,11 a “prohibitin domain” in the mid-region that is critical for lipid raft associations,11 a C-terminus containing a coiled-coil domain that is important for protein-protein interactions,11 and a small nuclear localization sequence that functions as a nuclear export signal.12 PHB1 translocates to the plasma membrane after undergoing certain PTMs.13 Also, different stimuli induce PHB1 nuclear-mitochondrial shuttling, like ethanol treatment in β-cells,14 estrogen in prostate cancer cells,15 and camptothecin in breast cancer cells.12 Nonetheless, the mechanisms regulating PHB1’s intracellular trafficking are not fully understood.
Mitochondrial PHB1 regulates mitochondrial turnover and apoptosis, nuclear PHB1 regulates transcription and cell cycle, and membrane-bound PHB1 regulates cell signaling, suggesting that PHB1 localization to a specific cellular compartment determines its function. PHB1 is also present in the circulation16 and can be internalized when added to cultured cells,17 which suggest that circulating PHB1 may also have some regulatory role. In this section, different biological functions of PHB1 in the mitochondria, nucleus, and plasma membrane are further discussed.
Mitochondrial PHB1
The best-described and least controversial function of PHB1 is as a mitochondrial chaperone. PHB1 heterodimerizes with its homolog PHB2 in the inner mitochondrial membrane to form a large multimeric complex, known as the PHB complex, to maintain the structure of mitochondria and regulate their function. The PHB complex protects newly imported proteins from being degraded by mitochondrial proteases, promotes mitochondrial protein synthesis, maintains the organization and copy number of mitochondrial DNA, and acts a chaperone for newly synthesized mitochondrial proteins, including subunits of the electron transport chain and the GTPase optic atrophy 1, which regulates cristae morphogenesis and mitochondrial dynamics.18 PHB1 is essential for normal mitochondrial development and function and its deficiency is associated with deficient mitochondrial biogenesis, abnormal cristae morphology, and reduced replicative lifespan in yeast and mammals.19 In view of the central role of PHB1 in maintaining normal structure and function of mitochondria, it is not surprising that its dysregulation has been associated with aging and a variety of proliferative, degenerative, metabolic, and inflammatory diseases.20,21
Multiple studies have shown that PHB1 also has a major role in cytoprotection by preventing oxidative stress and mitochondria-mediated apoptosis.1 Whereas the absence of PHB1 has been associated with increased generation of reactive oxygen species (ROS) and higher sensitivity to apoptotic stimuli, its overexpression prevented cell death in cardiomyocytes.22 Two mitochondrial-related mechanisms by which PHB1 exerts its protective role are: (1) through the inhibition of the intrinsic apoptotic pathway by maintaining the mitochondrial membrane potential and by inhibiting cytochrome c release and caspase-3 activation,2 and (2) through its interaction with the 66 kDa proto-oncogene Src homologous-collagen homologue (Shc) adaptor protein p66Shc, which is a major mediator of mitochondrial stress-induced apoptosis, as observed in a variety of cell types such as colonic and brain endothelial cells, granulosa cells, and cardiomyocytes.1 When p66Shc is activated by UVC or ROS, it induces the generation of mitochondrial ROS, triggering apoptosis.23 Interaction between PHB1 and p66Shc may block this response, although the precise mechanism remains to be defined.
Although there are few studies on the role of mitochondrial PHB1 in liver pathology, it is known to be essential for normal liver function. Methionine adenosyltransferase 1 A (Mat1a)-knockout (KO) mice have reduced PHB1 expression and impaired mitochondrial function, and spontaneously develop hepatocellular carcinoma (HCC).24 The liver specific Phb1-KO mouse model confirmed that reduced PHB1 expression contributes to the Mat1a-KO phenotype and similar to Mat1a-KO mice, these mice spontaneously develop liver injury and HCC.4 At only three weeks of age hepatic mitochondria of liver specific Phb1-KO mice appear swollen and many have no discernible cristae, and consistent with impaired mitochondrial function, liver specific Phb1-KO mice have increased lipid peroxidation and liver injury.4
Mitochondrial PHB1 exists as a heterodimer with PHB2. These two proteins are known to stabilize each other.25 Phb2-KO mice are, like Phb1-KO, lethal embryonically (http://www.informatics.jax.org/external/ko/lexicon/2210.html,19). Loss of PHB2 in mouse embryonic fibroblasts resulted in aberrant mitochondrial cristae morphogenesis and increased apoptosis,25 and it has been shown recently that liver specific Phb2-KO have mitochondrial fragmentation, increased apoptosis, and increased lipid accumulation.26 Whether these mice also spontaneously develop HCC will be of interest. Outside of the mitochondria, whether PHB1 also exists as a heterodimer with PHB2 is not clear.
Altogether, these findings demonstrate that mitochondrial PHB1, along with PHB2, is hepatoprotectant. Interestingly, although the role of mitochondrial PHB1 in liver malignant transformation has not been studied, it has been hypothesized in bladder cancer that it acts as an oncoprotein.27
Nuclear PHB1
PHB1 is also localized in the nucleus where it interacts, either directly or indirectly, with different transcription factors such as p53, retinoblastoma (Rb), E2F, apoptosis-inducing factor (AIF), c-MYC, c-FOS, and MYC-associated factor X (MAX), to regulate transcriptional activity.2 These tumor suppressors and oncogenes are major regulators of cell cycle and survival and may link the function of nuclear PHB1 to proliferation and apoptosis. In fact, it was reported that PHB1 nuclear accumulation can induce cell cycle arrest and inhibit proliferation.28 PHB1 overexpression inhibited growth of breast-cancer cells29 and androgen-dependent prostate cancer cells,30 among others.31 In 1999, Wang et al.32 demonstrated that PHB1 can act as a tumor suppressor by interacting physically with Rb family proteins and repressing E2F-mediated transcription. Later, they observed that PHB1-mediated transcriptional repression required histone-deacetylase (HDAC) activity and co-repressors like the nuclear receptor co-repressor (N-CoR).33 Fusaro et al.34 demonstrated that PHB1 can also interact physically with p53 to enhance its transcriptional activity. In this study, the authors showed that PHB1 induces p53-mediated transcription by enhancing its recruitment to promoter regions. However, Jupe et al.,35 showed that the anti-proliferative activity of PHB1 resides, at least in part, in its 3ʹUTR region. In their study, mutations in the 3′UTR were identified in multiple human cancer cell lines including cervical carcinoma, glioblastoma, bladder carcinoma, and transformed skin fibroblasts, in which the delivery of PHB1 3′UTR arrested the cell cycle.35 Years later, the same group demonstrated that PHB1 3ʹUTR produces a functional RNA that functions as a tumor suppressor in human breast cancer.36 In the last few years, our group has identified additional mechanisms by which nuclear PHB1 regulates gene expression that result in cytoprotection and tumor suppression in the liver. We reported that PHB1 can directly interact with nuclear erythroid 2 p45-related factor 2 (NRF2). Through this interaction, PHB1 serves as a co-activator of the antioxidant response element (ARE) to positively regulate ARE-dependent genes and provide cellular protection.37 In terms of tumor suppressive mechanisms, PHB1 acts as a co-repressor with the CCCT-binding factor (CTCF) on the imprinting control region (ICR) to suppress the expression of insulin-like growth factor 2 (IGF2) and H19 and inhibit hepatocyte proliferation.38 Furthermore, PHB1 heterodimerizes with MAX to bind and repress E-box promoter activity, suppressing c-MYC, MAFG, and c-MAF expression in HCC and cholangiocarcinoma (CCA) cells.7 PHB1 also acts as a negative regulator of the Wingless-related integration site (WNT)/β-catenin signaling in the liver, in part through E2F1.8 Finally, we recently showed that reduced PHB1 expression induced interleukin-8 (IL-8) transcription by activating nuclear factor-kappa B (NF-κB) and activator protein 1 (AP-1), which resulted in enhanced IL-8 expression and release to promote tumorigenesis.9 PHB1 was unable to bind to the IL-8 promoter by itself but it was able to bind in the presence of c-JUN, a finding that suggests the possibility of PHB1 acting as a co-repressor of the AP-1 site.9
Plasma membrane-associated PHB1
PHB1 also targets lipid rafts and has been detected in the plasma membrane where it acts as a transmembrane adaptor to activate downstream signaling pathways.1 In multiple cancer cells like cervical and pancreatic cancer cells, membrane-bound PHB1 is required for the association of c-RAF to the plasma membrane and the activation of RAS-mediated RAF-mitogen-activated protein kinase kinase (MEK)-extracellular receptor kinase (ERK) signaling.39,40 This mechanism modulates cancer cell survival and migration, supporting the role of PHB1 as an oncoprotein. Conversely, a study by Pan et al.41 identified membrane-associated PHB1 as a pro-apoptosis mediator in liver fibrosis. Pan et al.41 demonstrated that Tan IIA – a diterpene quinine derivative – treatment led to a predominant membrane localization of PHB1 and the translocation of c-RAF from the cytoplasm to the membrane in hepatic stellate cells (HSCs). PHB1 silencing and loss of membrane-bound PHB1 disrupted the PHB1/c-RAF complex, attenuating Tan IIA-induced apoptosis of activated HSCs. It should be noted that depletion of HSCs by apoptosis is a potential strategy for the treatment of liver fibrosis. Regulatory mechanisms for PHB1 plasma membrane targeting and the reasons for the opposing effects of plasma membrane-associated PHB1 in different cell types are largely unknown.
Membrane-bound PHB1 can act as a receptor for certain bacteria and viruses. For instance, PHB1 was shown to bind to the Vi capsular polysaccharide of Salmonella typhi in human intestinal epithelial cells.42 PHB1 has also been shown to function as a receptor for Chikungunya virus in microglial cells43 and dengue serotype 2 virus.44 In the liver, hepatocyte membrane-bound PHB1 facilitates the entry of hepatitis C virus (HCV).45
Lastly, cell membrane-associated PHB1 also acts as specific receptor in white fat blood vessels.46 PHB1 is highly expressed in the endothelial cells of white adipose tissue in both mice and humans and plasma membrane-bound PHB1 mediates the internalization of an adipose-specific peptide.46 More importantly, a specific PHB1 receptor complex in the white adipose vasculature has been shown to contribute to the induction of apoptosis in the endothelial cells of white fat blood vessels, preventing diet-induced obesity.47
Besides the studies on HCV and HSC, the functional role of membrane bound PHB1 in the liver is largely unknown.
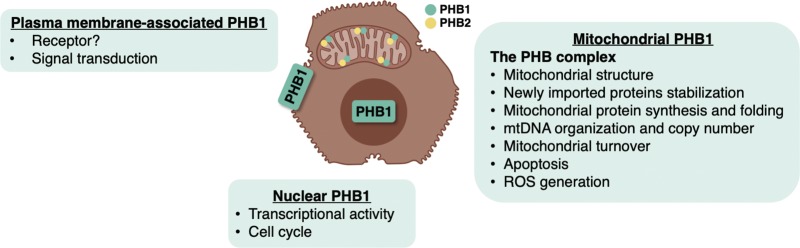
Figure 1 summarizes the PHB1’s roles in different cellular compartments.
Figure 1.
PHB1’s roles in different cellular compartments. PHB1 can exert different functions depending on its subcellular localization. Plasma membrane-bound PHB1 activates multiple signaling pathways and has been shown to function as a receptor for some viruses and bacteria in other cell types. In hepatocytes it has been shown to facilitate HCV entry. Whether hepatocyte membrane-bound PHB1 also binds to other viruses or bacteria is unknown. Nuclear PHB1 regulates transcription and cell proliferation by interacting with different transcription factors involved in the cell cycle. In the mitochondria, PHB1 interacts with PHB2 to form the PHB complex and regulate processes essential for normal mitochondrial function. (A color version of this figure is available in the online journal.)
Regulation of PHB1 expression
Although numerous publications have documented roles of PHB1 in different diseases, little is known about how its expression is regulated. Knowing how a gene or protein is regulated is crucial not only to understand its functions in both health and disease, but also to facilitate the design of new drugs and therapies that allow its targeting. In this section, we will summarize the known mechanisms that regulate PHB1 at the transcriptional, post-transcriptional, and post-translational levels.
Transcriptional regulation
The transcriptional regulation of PHB1 is not completely understood. Increased expression of PHB1 in certain cancers has been attributed to c-MYC binding sites in its promoter, suggesting that c-MYC activates PHB1 transcriptionally.48 However, proof of the ability of c-MYC to trans-activate PHB1 is lacking. On the contrary, we found that c-MYC suppresses PHB1 expression in liver cancer cells through a mechanism involving a repressive E-box at −256 of the PHB1 gene.7 Interestingly, while the repressive E-box element in PHB1 is occupied by MAX, methionine adenosyltransferase α1 (MATα1), PHB1, and Max’s Next Tango (MNT) in normal hepatocytes and cholangiocytes, it switched to MAX, c-MYC, c-MAF, and MAFG in liver cancer cells.7 These proteins exert their influence on PHB1 transcription via this E-box so that the wild-type (but not E-box mutant) PHB1 promoter is positively regulated by MATα1 but negatively regulated by c-MYC, MAFG, and c-MAF.7 A study performed by Puppin et al.49 demonstrated that PHB1 is also regulated by histone acetylation.49 Treatment of thyroid tumor cells with the HDAC inhibitors trichostatin A and sodium butyrate significantly increased PHB1 expression and confirmed that HDAC1 and HDAC2 regulate PHB1 transcription and alternative splicing.49 Finally, Theiss et al.50 showed that interleukin-6 (IL-6) transcriptionally regulates PHB1 in intestinal epithelial cells. IL-6 induces STAT3 binding to the IL-6 response element in the PHB1 promoter, thereby inducing PHB1 promoter activity and increasing PHB1 protein and mRNA levels. This regulation could also explain PHB1 upregulation in certain HCCs, as IL-6 signaling is important in the initiation and development of HCC.51
Post-transcriptional regulation
Multiple miRNAs bind to the 3ʹ-UTR of PHB1 destabilizing the mRNA and leading to reduced PHB1 expression. miRNA-26a in glioma cells52 and miRNA-27a in prostate cancer,53 gastric adenocarcinoma,54 and glioma cells55 have been shown to promote tumorigenesis by targeting PHB1 mRNA 3ʹ-UTR. On the other hand, miRNA-195 has recently been described as anti-proliferative in human melanoma cells by also targeting PHB1,56 and miRNA-128 has been found to suppress PHB1 expression and promote apoptosis in cardiomyocytes.57 In the liver, our group found that c-MYC induces the expression of miRNA-27a/b, which also target PHB1 mRNA 3ʹ-UTR and lower its expression.37 Lastly, a recent study confirmed that the long non-coding RNA PHBP1 (prohibitin gene pseudogene 1) promotes the stabilization of PHB1 mRNA.58
Post-translational regulation
PHB1 PTMs influence its subcellular localization and activity and are hence critical for downstream effects on proliferation and survival. PHB1 undergoes several PTMs including phosphorylation,59–61 O-linked beta-N-acetylglucosamine (O-GlcNAc) modification,61 palmitoylation,13 ubiquitination,62 and cysteine oxidation.63 Phosphorylation is a key modification that regulates PHB1 function, intracellular trafficking, and protein binding specificity. PHB1 can be phosphorylated at several serine/threonine and tyrosine residues and phosphorylation at different sites has opposite effects on tumorigenesis. For instance, while AKT phosphorylation of PHB1 at Thr258 inhibits its interaction with the Src homology region 2 domain-containing phosphatase-1 (SHP1), thereby activating the phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathway,64 PHB1 phosphorylation at Tyr114 facilitates its interaction SHP1, blocking PI3K/AKT signaling.60 The phosphorylation of PHB1 at Thr258 in the plasma membrane of cancer cells is an important event in PHB1 biology since it leads to the activation of both the c-RAF/MEK/ERK and PI3K/AKT signaling pathways, favoring proliferation and metastasis.65
Considering that PHB1’s different biological functions are mainly attributed to its subcellular localization, uncovering the mechanisms regulating its subcellular trafficking has been of major interest. While PHB1 mitochondrial targeting is driven by AKT phosphorylation at Thr258,27 PHB1 palmitoylation at Cys69 facilitates its translocation to the membrane where it can undergo tyrosine phosphorylation.13 It should be noted that these modifications can occur in response to stimuli like insulin and growth factors or under stress and pathological conditions, suggesting that PHB1 functions are important in both health and disease.66
PHB1 PTMs have not been studied in the liver so how they regulate PHB1 function is unknown.
PHB1 in liver injury
In the majority of the studies, PHB1 has been found to be tumor suppressive and hepatoprotective in the liver. By multiple mechanisms that will be discussed in this section, low PHB1 expression predisposes the liver to injury and the development of HCC and CCA, the two most common primary liver cancers.
As previously mentioned, a proteomics study by Santamaria et al.67 observed that mice lacking MAT1A, which spontaneously develop steatohepatitis (NASH) and HCC and are characterized by mitochondrial dysfunction, have reduced PHB1 expression. Interestingly, lower PHB1 protein levels were also found in livers from ob/ob mice and obese patients at high risk for NASH.67 These findings suggested a protective role for PHB1 in the liver and led to the generation of a liver-specific Phb1-KO mouse model4 that has allowed us to study the function of PHB1 in liver physiology and disease in vivo and to identify new mechanisms of PHB1 regulation. A tissue-specific deletion was necessary because total body deletion of Phb1 is embryonically lethal [Lexicon Knockout Mice Phenotype Data Summary NIH-1165].
The liver-specific Phb1-KO mouse model
PHB1 has a protective role in the liver as liver-specific Phb1-KO mice exhibit severe liver injury at only three weeks old.4 Liver-specific Phb1-KO mice have increased oxidative stress, apoptosis, and fibrosis and as already discussed, abnormal mitochondria.4 Liver-specific Phb1-KO liver’s mitochondria are swollen and the majority have no discernible cristae. In addition, hepatocyte dysplasia, bile duct epithelial metaplasia, increased proliferation and staining for stem cell and preneoplastic markers were observed in the absence of Phb1.4 From 20 weeks on, liver-specific Phb1-KO mice have multiple liver nodules and by eight months, around 40% developed multifocal HCC.4
To gain more insight into PHB1’s protective role in the liver, a microarray analysis of 25,000 genes examined differential mRNA expression profiles in liver-specific Phb1-KO and Phb1-Flox livers. The analysis revealed up-regulation of 402 genes, many of them associated with liver fibrosis, growth, and tumorigenesis, suggesting that PHB1 is involved in liver malignant transformation. Other differentially expressed genes were associated with processes such as inflammation and angiogenesis, supporting a major role of PHB1 in liver pathology. Since liver-specific Phb1-KO mice have liver injury, development of HCC could have been due to chronic inflammation. However, PHB1 exerted an anti-proliferative effect in AML-12 (alpha mouse liver 12) cells, a murine hepatocyte cell line, and its knockdown raised cyclin D1 and H19 expression and cell growth.4 Our subsequent investigations further strengthened the notion that PHB1 acts mainly as a tumor suppressor in HCC and CCA (see below).
PHB1 in cholestatic liver injury
We have shown that PHB1 expression is downregulated at the mRNA and protein levels in human cholestatic liver diseases and mouse models of chronic cholestatic liver injury.6,37 Treatment with the toxic bile acid lithocholic acid (LCA) lowered PHB1 expression in vivo. Similarly, bile duct ligation (BDL), a commonly used murine model for cholestatic liver disease, also lowered PHB1 mRNA and protein levels.6,37 As mentioned above, liver-specific Phb1-KO mice have bile duct epithelial metaplasia at a very early age, suggesting that PHB1 deficiency predisposes to bile duct abnormality. Liver-specific Phb1-KO mice developed more injury after BDL,6 and Phb1 heterozygotes developed aberrant bile duct proliferation (one developed CCA) after left and median BDL,7 demonstrating that the loss of Phb1 sensitizes to cholestatic liver injury and subsequent malignant transformation. Finally, confirming the finding in humans, PHB1 mRNA levels were lower in patients with primary biliary cholangitis (PBC), biliary atresia (BA), and Alagille syndrome (ALS), three common cholestatic conditions in adults (PBC) and children (BA and ALS).6
Although most of the work regarding PHB1 in liver injury has been done on cholestatic liver disease, PHB1 might be also involved in non-alcoholic fatty liver disease and acute liver inflammation. Phb1 heterozygotes had more liver steatosis after being fed with a methionine and choline deficient diet and higher mortality and liver injury after being challenged with bacterial lipopolysaccharide and concanavalin A, two-classical pro-inflammatory insults.5
Mechanisms of PHB1’s cytoprotection
In recent years, both PHB1 mitochondrial-dependent and -independent functions for cell protection have been elucidated. Mitochondrial-dependent functions of PHB1 have been summarized in the ‘Mitochondrial PHB1’ section. In this section, we summarize mitochondrial-independent mechanisms known to date by which PHB1 protects the liver from injury.
NRF2 activation
PHB1 interacts with NRF2 directly to protect the liver from injury.37 NRF2 represents a major mechanism for cellular defense against oxidative stress as the activation of the NRF2-ARE signaling pathway induces the expression of genes involved in detoxication and elimination of reactive oxidants. Yang et al.37 reported that the activation of c-MYC via miRNA27a/b in both in vitro (LCA treatment) and in vivo (BDL and LCA treatment) models of cholestasis decreased the expression of PHB1 and NRF2, suppressing the expression of NRF2-ARE target genes such as glutamate-cysteine ligase (GCL) subunits, thereby lowering glutathione (GSH) level and the cellular antioxidant capacity. Interestingly, both c-MYC and PHB1 interact with NRF2 and influence its trans-activating activity at the ARE. However, they do so in an opposite manner; c-MYC is a co-repressor, whereas PHB1 is a co-activator of NRF2 at the ARE. While silencing of either c-MYC or miR27a/b attenuated LCA-induced downregulation of NRF2, PHB1, and GCL, PHB1 overexpression protected against the suppression of NRF2 and GCL expression.37 These findings provided a novel and key mechanism of how PHB1 deficiency may predispose to chronic cholestatic injury.
Influence on MAT1A
We recently showed a positive regulation between PHB1 and MAT1A.7 MAT1A encodes for MATα1, the enzyme that catalyzes the synthesis of S-adenosylmethionine (SAMe) in the liver.68 SAMe is the main methyl donor of the cell and is used for the methylation of biomolecules such as DNA, RNA, proteins, and phospholipids.68 Besides methylation, SAMe is a precursor of GSH and polyamine synthesis and hence, any variation on its metabolism may have a major impact on cell growth, differentiation, and survival.69 MAT1A is highly expressed in normal liver and specifically in hepatocytes and bile duct epithelial cells; like PHB1, it is repressed during liver injury and liver cancer.6,7,70 We found that PHB1 and MATα1 interact directly in normal liver and that together, they act as tumor suppressors. While their overexpression inhibited growth in HCC and CCA cells, their knockdown increased it. Overexpression of PHB1 enhanced MAT1A mRNA and protein levels and reduced the expression of the oncogenes c-MYC/MAFG/c-MAF in both HCC and CCA cells, and PHB1 silencing had the opposite effect.7 Thus, PHB1 can protect the liver from injury and malignant transformation via enhancing MAT1A expression.
HDAC4 subcellular localization
PHB1 also influences HDAC4 expression and activity.6 HDAC4 is a type II histone deacetylase that shuttles between cytosol and nucleus to regulate gene expression. HDAC4 is upregulated in liver fibrosis and liver cancer, and a greater nuclear localization of HDAC4 has been attributed to lower PHB1 levels in cholestatic liver disease.6 As seen in liver-specific Phb1-KO and wild-type mice after Phb1 silencing, low PHB1 expression increased HDAC4 expression and nuclear content in the liver, which was believed to enhance the apoptotic response to bile acids in hepatocytes.6 Hdac4 silencing or its inhibition using the HDAC inhibitor parthenolide protected liver-specific Phb1-KO mice from BDL-induced cholestatic injury.6 HDAC4 inhibition restored the expression of genes associated with bile acid metabolism and inflammation that were deregulated in liver-specific Phb1-KO mice, supporting an epigenetic deregulation caused by nuclear accumulation of HDAC4 in the context of Phb1 deficiency.
Activation of inflammatory cells
Reduced PHB1 level has been associated with inflammatory pathologies.20,21 At only three weeks old, liver-specific Phb1-KO mice have inflammation of the liver, as observed histologically and by the upregulation of multiple inflammatory genes.4 Sánchez-Quiles et al.5 showed that Phb1 deficiency also leads to the activation of lymphocytic cells, enhancing the inflammatory response and the sensitivity to inflammatory insults in the liver.5 They found that the proliferation of splenocytes in response to anti-CD3 (cluster of differentiation 3) and the expression of mediators of lymphoid cell activation such as PP2A (protein phosphatase 2 A), ERK, AKT, and glycogen synthase kinase 3 beta (GSK3β) were significantly higher in Phb1 heterozygotes.5 Furthermore, transcriptomics performed in livers from wild-type and Phb1 heterozygotes showed deregulation of genes involved in inflammation and cancer. Lastly, circulating inflammatory cytokines were increased in serum from Phb1 heterozygotes, strongly suggesting that Phb1 deficiency is associated with an inflammatory condition that may contribute to the progression of liver injury.5
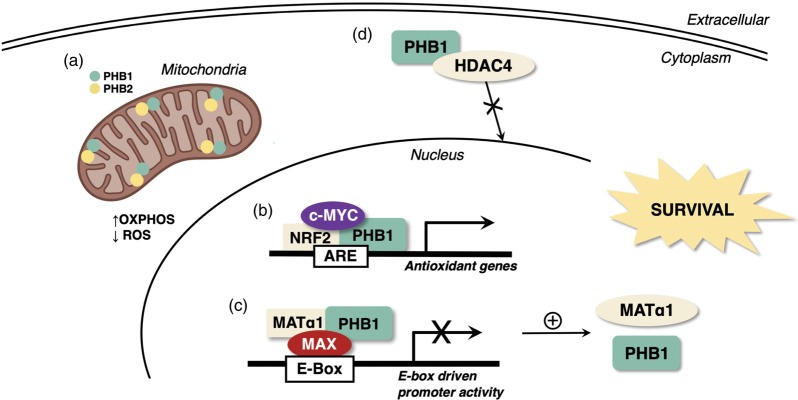
Figure 2 summarizes how PHB1 protects the liver against injury.
Figure 2.
How PHB1 protects the liver against injury. There are multiple mechanisms by which PHB1 protects the liver against injury. (a) Mitochondrial PHB1 maintains mitochondrial respiration and limits the generation of ROS, preventing apoptosis. In the nucleus, (b) PHB1 interacts with NRF2 to transactivate the antioxidant response element (ARE), which is present in the promoter region of many genes involved in antioxidant cellular defense, and (c) heterodimerizes with MAX and MATα1 to repress E-box-driven transcription, which suppresses several oncogenes but positively regulates PHB1 and MATα1 expression since they both have repressor E-boxes. (d) In the cytosol, PHB1 interacts with HDAC4 to prevent its nuclear translocation and associated epigenetic changes. (A color version of this figure is available in the online journal.)
PHB1 in liver cancers
As mentioned earlier, the most controversial aspect of PHB1 is in cancer as it can exert both pro- and anti-tumorigenic effects even in the same organ, as seen in breast and prostate cancers.71,72 Although a similar conflict exists for PHB1 in HCC as well,7,73 most of the publications point to its role as a tumor suppressor. For this reason, the underlying mechanisms for PHB1 tumor suppressive function have been better characterized and in this review we focus on its tumor suppressive role in the liver.
H19/IGF2 axis
One way that PHB1 controls cell growth in hepatocytes is by regulating the H19- IGF2 axis. H19 and IGF2, which are associated with tumor growth, were first found upregulated in the livers of three-week-old liver-specific Phb1-KO mice.4 Phb1 silencing in the murine hepatocyte cell line AML-12 and the mouse HCC cell line SAMe-D, as well as in the human liver cancer cell lines HepG2 and Huh7 confirmed that PHB1 negatively regulates H19 and IGF2.38 Both H19 and IGF2 are regulated by CTCF and co-immunoprecipitation analysis revealed that CTCF and PHB1 bind to the ICR element of H19 and IGF2 to repress their expression. Notably, human HCC samples with high H19 and IGF2 had reduced PHB1 and CTCF expression and ICR binding activity.38 Interestingly, Phb1 silencing lowered CTCF protein levels and CTCF ICR binding activity, and CTCF overexpression reduced H19 and IGF2 expression levels and also prevented their induction after Phb1 knockdown.38 Furthermore, silencing Phb1 or overexpressing H19 enhanced cell growth in SAMe-D cells. While overexpressing H19 enhanced Phb1 knockdown-mediated cell growth, its inhibition had the opposite effect, suggesting that the effect of Phb1 on cell growth and tumorigenesis occurs at least partially via H19. Interestingly there is a reciprocal negative regulation between H19 and PHB1, as silencing H19 raised PHB1 expression.38 How H19 regulates PHB1 expression and whether PHB1 is an important target of H19 remains to be examined. Taken together, these results demonstrated that PHB1 and CTCF cooperate to negatively regulate the H19-IGF2 axis and cell growth in hepatocytes.38
β-catenin/WNT pathway
Another mechanism by which PHB1 acts as a tumor suppressor in the liver is by suppressing the WNT/β-catenin signaling pathway.8 This pathway is implicated in liver development and regeneration74,75 and its overactivation has been correlated with HCC in both human and mice.75 Liver-specific Phb1-KO mice have increased hepatic expression of WNT target genes and hyperactivation of GSK3β and AKT, as determined by the phosphorylation of GSK3β-Ser9 and AKT-Ser473, respectively.8 Multiple WNT ligands including Wnt7a and Wnt10a are upregulated at mRNA level in liver-specific Phb1-KO livers and while PHB1 knockdown activated WNT signaling in HepG2 cells, its overexpression inhibited it.8 Interestingly, PHB1 suppressed multiple WNT ligands in an E2F1-dependent manner. Chromatin immunoprecipitation analyses showed increased binding of E2F1 to the Wnt10a promoter in liver-specific Phb1-KO livers and WNT9A in HepG2 cells after PHB1 silencing.8 Taken together, PHB1 deletion activates the WNT/β-catenin signaling pathway by releasing the inhibition that it exerts on the expression of multiple WNT ligands.
c-MYC/MAFG/c-MAF suppression
During cholestatic liver injury, PHB1 expression falls but the expression of c-MYC, MAFG, and c-MAF is induced.7,70 This finding prompted us to examine the interplay between PHB1 and c-MYC/MAFG/c-MAF and we uncovered a reciprocal negative regulation between PHB1 and c-MYC/MAFG/c-MAF. PHB1 heterodimerizes with MAX and binds to the E-box element to repress E-box-dependent promoter activity.7 Interestingly, c-MYC, MAFG, and c-MAF all have enhancer E-box elements in their promoter regions, whereas PHB1 promoter contains a repressive E-box element.7 Consistently, PHB1 overexpression suppressed the expression of c-MYC/MAFG/c-MAF while PHB1 silencing had the opposite effect.7 Since PHB1 positively regulates MAT1A, which also suppresses the expression of c-MYC/MAFG/c-MAF, we evaluated whether PHB1’s effect requires MAT1A using SAMe-D cells. We found that PHB1 still suppressed the expression of c-MYC/MAFG/c-MAF, demonstrating that it is able to regulate their expression independent of MAT1A.7
NF-κB pathway
PHB1 was shown to modulate NF-κB signaling.76 PHB1 expression is lower in patients with inflammatory bowel disease (IBD) and Theiss et al.76 demonstrated that PHB1 inhibits tumor necrosis factor alpha (TNFα)-induced nuclear translocation of NF-κB in the intestine. TNFα and NF-κB play major roles in the pathogenesis of IBD and also in the development of HCC.77 Theiss et al. found a crosstalk between PHB1 and TNFα, while TNFα reduces PHB1 expression at protein and mRNA levels, sustained expression of PHB1 decreases TNFα-induced NF-κB activity. Specifically, PHB1 inhibited p65 (subunit of NF-κB) nuclear translocation, the binding of NF-κB to DNA and as a consequence, NF-κB-mediated transcription. Importin α3 is involved in the nuclear translocation of NF-κB and its expression was reduced when PHB1 was overexpressed.76 Interestingly, maintaining high levels of importin α3 after PHB1 overexpression sustained the activation of NF-κB by TNF-α suggesting that PHB1 inhibits NF-κB nuclear translocation by regulating importin α3 expression level.76 Thus, PHB1 protects intestinal epithelial cells from the deleterious effects of TNFα-and NF-κB. We also found that increased p65 nuclear translocation occurred in liver cancer cells after PHB1 silencing,9 supporting the notion that this could be another tumor suppressive mechanism of PHB1 in the liver.
IL-8 levels
Increased expression of the pro-inflammatory CXC chemokine IL-8 has been reported in liver cancer cells78 and serum of patients with HCC.79 IL-8 activates multiple signaling pathways and has a major role in the tumor microenvironment. Indeed, IL-8 produced by liver cancer cells has angiogenic activity80 and acts as an autocrine growth factor.81 Using RNA-sequencing analysis, we found that IL-8 is negatively regulated by PHB1 in HCC and CCA cells.9 PHB1 silencing increased IL-8 at both protein and mRNA levels, while PHB1 overexpression significantly repressed it. Supporting this regulation, there is an inverse correlation between PHB1 and IL-8 mRNA levels in HCCs with reduced PHB1 expression and higher IL-8 levels, correlating with a poorer prognosis in HCC patients.9 c-JUN N-terminal kinase (JNK) and NF-κB signaling pathways were found to be required for PHB1-silencing induced IL-8 expression, as the specific inhibition of both NF-κB and JNK blocked IL-8 upregulation. Furthermore, PHB1 knockdown increased IL-8 promoter activity through AP-1 and NF-κB by increasing the nuclear content of p65 and c-JUN and their binding to the IL-8 promoter. It should be noted that PHB1 is not able to bind to the DNA by itself but it is able in the presence of c-JUN.9 Finally, PHB1 knockdown induced IL-8 secretion, migration, and invasion in liver cancer cells.9 Addition of conditioned media from PHB1 silenced HepG2 cells to parental HepG2 or SK-Hep cells increased migration and invasion, which was blocked by neutralizaing antibody to IL-8.9 This mechanism describes how low PHB1 expression in liver cancer can promote a more aggressive phenotype.
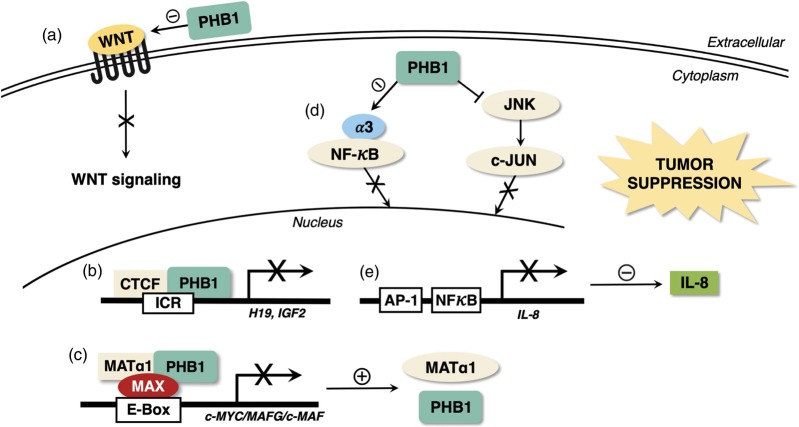
Figure 3 summarizes how PHB1 exerts tumor suppression in the liver.
Figure 3.
How PHB1 exerts tumor suppression in the liver. There are several mechanisms by which PHB1 acts as a tumor suppressor in the liver. (a) PHB1 negatively regulates the expression of multiple WNT ligands, thereby inhibiting the activation of the canonical WNT signaling pathway. (b) In the nucleus, PHB1 and CTCF cooperate to negatively regulate the H19-Igf2 axis and inhibit cell proliferation. (c) Nuclear PHB1 heterodimerizes with MAX and MATα1 to repress the expression of the oncogenes c-MYC, MAFG, and c-MAF and enhance the expression of PHB1 itself and MATα1, which also acts as a tumor suppressor. (d) PHB1 reduces the expression of importin α3, inhibiting NF-κB nuclear translocation. (e) PHB1 reduces IL-8 transcription and release by lowering the nuclear content of NF-κB and c-JUN, thus inhibiting cell invasion and migration. (A color version of this figure is available in the online journal.)
Conclusions and future directions
Over the past few years, PHB1 has emerged as a key player that maintains liver health. In addition to its well-known role as a mitochondrial chaperone, it is increasingly clear that PHB1 has cytoprotective function and anti-tumor activity in the liver that are independent of the mitochondria. The most perplexing aspect of PHB1 biology is in cancer, as it can be either tumor suppressive or oncogenic even in the liver. It is likely that the subcellular localization, PTMs, and interacting proteins are major determinants in how PHB1 behaves. Future research should better identify these mechanisms, which may lead to novel therapeutic strategies targeting PHB1.
Authors’ contributions
All authors contributed to this paper with literature review and analysis, drafting and critical revision and editing, and final approval of the submitted version.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the NIH [grant number R01CA172086] (to Shelly C. Lu). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ORCID iD
Shelly C Lu https://orcid.org/0000-0003-2128-5407
References
- 1.Thuaud F, Ribeiro N, Nebigil CG, Désaubry L. Prohibitin ligands in cell death and survival: mode of action and therapeutic potential. Chem Biol 2013; 20:316–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng YT, Chen P, Ouyang RY, Song L. Multifaceted role of prohibitin in cell survival and apoptosis. Apoptosis Int Apoptosis 2015; 20:1135–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McClung JK, Danner DB, Stewart DA, Smith JR, Schneider EL, Lumpkin CK, Dell’Orco RT, Nuell MJ. Isolation of a cDNA that hybrid selects antiproliferative mRNA from rat liver. Biochem Biophys Res Commun 1989; 164:1316–22 [DOI] [PubMed] [Google Scholar]
- 4.Ko KS, Tomasi ML, Iglesias-Ara A, French BA, French SW, Ramani K, Lozano JJ, Oh P, He L, Stiles BL, Li TWH, Yang H, Martínez-Chantar ML, Mato JM, Lu SC. Liver-specific deletion of prohibitin 1 results in spontaneous liver injury, fibrosis, and hepatocellular carcinoma in mice. Hepatology 2010; 52:2096–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sánchez-Quiles V, Segura V, Bigaud E, He B, O’Malley BW, Santamaría E, Prieto J, Corrales FJ. Prohibitin-1 deficiency promotes inflammation and increases sensitivity to liver injury. J Proteomics 2012; 75:5783–92 [DOI] [PubMed] [Google Scholar]
- 6.Barbier-Torres L, Beraza N, Fernández-Tussy P, Lopitz-Otsoa F, Fernández-Ramos D, Zubiete-Franco I, Varela-Rey M, Delgado TC, Gutiérrez V, Anguita J, Pares A, Banales JM, Villa E, Caballería J, Alvarez L, Lu SC, Mato JM, Martínez-Chantar ML. Histone deacetylase 4 promotes cholestatic liver injury in the absence of prohibitin-1. Hepatology 2015; 62:1237–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan W, Yang H, Liu T, Wang J, Li TWH, Mavila N, Tang Y, Yang J, Peng H, Tu J, Annamalai A, Noureddin M, Krishnan A, Gores GJ, Martínez-Chantar ML, Mato JM, Lu SC. Prohibitin 1 suppresses liver cancer tumorigenesis in mice and human hepatocellular and cholangiocarcinoma cells. Hepatology 2017; 65:1249–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mavila N, Tang Y, Berlind J, Ramani K, Wang J, Mato JM, Lu SC. Prohibitin 1 acts as a negative regulator of wingless/Integrated-Beta-Catenin signaling in murine liver and human liver cancer cells. Hepatol Commun 2018; 2:1583–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang JW, Murray B, Barbier-Torres L, Liu T, Liu Z, Yang H, Fan W, Wang J, Li Y, Seki E, Mato JM, Lu SC. The mitochondrial chaperone prohibitin 1 negatively regulates interleukin-8 in human liver cancers. J Biol Chem 2019; 294:1984–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mishra S, Murphy LC, Murphy LJ. The prohibitins: emerging roles in diverse functions. J Cell Mol Med 2006; 10:353–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter A, Kämäräinen O, Hofmann A. Molecular modeling of prohibitin domains. Proteins 2007; 68:353–62 [DOI] [PubMed] [Google Scholar]
- 12.Rastogi S, Joshi B, Fusaro G, Chellappan S. Camptothecin induces nuclear export of prohibitin preferentially in transformed cells through a CRM-1-dependent mechanism. J Biol Chem 2006; 281:2951–9 [DOI] [PubMed] [Google Scholar]
- 13.Ande SR, Mishra S. Palmitoylation of prohibitin at cysteine 69 facilitates its membrane translocation and interaction with eps 15 homology domain protein 2 (EHD2). Biochem Cell Biol Biochim Biol 2010; 88:553–8 [DOI] [PubMed] [Google Scholar]
- 14.Lee JH, Nguyen KH, Mishra S, Nyomba B. Prohibitin is expressed in pancreatic beta-cells and protects against oxidative and proapoptotic effects of ethanol. FEBS J 2010; 277:488–500 [DOI] [PubMed] [Google Scholar]
- 15.Dong P, Jiang L, Liu J, Wu Z, Guo S, Zhang Z, Zhou F, Liu Z. Induction of paclitaxel resistance by ERα mediated prohibitin mitochondrial-nuclear shuttling. PLoS One 2013; 8:e83519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mengwasser J, Piau A, Schlag P, Sleeman JP. Differential immunization identifies PHB1/PHB2 as blood-borne tumor antigens. Oncogene 2004; 23:7430–5 [DOI] [PubMed] [Google Scholar]
- 17.Vessal M, Mishra S, Moulik S, Murphy LJ. Prohibitin attenuates insulin-stimulated glucose and fatty acid oxidation in adipose tissue by inhibition of pyruvate carboxylase. FEBS J 2006; 273:568–76 [DOI] [PubMed] [Google Scholar]
- 18.Nijtmans LG, de Jong L, Artal Sanz M, Coates PJ, Berden JA, Back JW, Muijsers AO, van der Spek H, Grivell LA. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J 2000; 19:2444–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coates PJ, Jamieson DJ, Smart K, Prescott AR, Hall PA. The prohibitin family of mitochondrial proteins regulate replicative lifespan. Curr Biol CB 1997; 7:607–10 [DOI] [PubMed] [Google Scholar]
- 20.Nijtmans LGJ, Artal SM, Grivell LA, Coates PJ. The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell Mol Life Sci CMLS 2002; 59:143–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theiss AL, Sitaraman SV. The role and therapeutic potential of prohibitin in disease. Biochim Biophys Acta 2011; 1813:1137–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Ren Z, Zhan R, Wang X, Wang X, Zhang Z, Leng X, Yang Z, Qian L. Prohibitin protects against oxidative stress-induced cell injury in cultured neonatal cardiomyocyte. Cell Stress Chaperones 2009; 14:311–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gertz M, Fischer F, Wolters D, Steegborn C. Activation of the lifespan regulator p66Shc through reversible disulfide bond formation. Proc Natl Acad Sci USA 2008; 105:5705–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martínez-Chantar ML, Corrales FJ, Martínez-Cruz LA, García-Trevijano ER, Huang Z-Z, Chen L, Kanel G, Avila MA, Mato JM, Lu SC. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J 2002; 16:1292–4 [DOI] [PubMed] [Google Scholar]
- 25.Merkwirth C, Dargazanli S, Tatsuta T, Geimer S, Löwer B, Wunderlich FT, von Kleist-Retzow JC, Waisman A, Westermann B, Langer T. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev 2008; 22:476–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Martin-Levilain J, Jiménez-Sánchez C, Karaca M, Foti M, Martinou JC, Maechler P. In vivo stabilization of OPA1 in hepatocytes potentiates mitochondrial respiration and gluconeogenesis in a prohibitin-dependent way. J Biol Chem 2019; 294:12581–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang L, Dong P, Zhang Z, Li C, Li Y, Liao Y, Li X, Wu Z, Guo S, Mai S, Xie D, Liu Z, Zhou F. Akt phosphorylates prohibitin 1 to mediate its mitochondrial localization and promote proliferation of bladder cancer cells. Cell Death Dis 2015; 6:e1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu YH, Peck K, Lin JY. Involvement of prohibitin upregulation in Abrin-triggered apoptosis. Evid Based Complement Alternat Med 2012; 2012:605154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng X, Mehta R, Wang S, Chellappan S, Mehta RG. Prohibitin is a novel target gene of vitamin D involved in its antiproliferative action in breast cancer cells. Cancer Res 2006; 66:7361–9 [DOI] [PubMed] [Google Scholar]
- 30.Dart DA, Spencer-Dene B, Gamble SC, Waxman J, Bevan CL. Manipulating prohibitin levels provides evidence for an in vivo role in androgen regulation of prostate tumours. Endocr Relat Cancer 2009; 16:1157–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra S, Murphy LC, Nyomba BLG, Murphy LJ. Prohibitin: a potential target for new therapeutics. Trends Mol Med 2005; 11:192–7 [DOI] [PubMed] [Google Scholar]
- 32.Wang S, Nath N, Adlam M, Chellappan S. Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene 1999; 18:3501–10 [DOI] [PubMed] [Google Scholar]
- 33.Wang S, Fusaro G, Padmanabhan J, Chellappan SP. Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene 2002; 21:8388–96 [DOI] [PubMed] [Google Scholar]
- 34.Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem 2003; 278:47853–61 [DOI] [PubMed] [Google Scholar]
- 35.Jupe ER, Liu XT, Kiehlbauch JL, McClung JK, Dell’Orco RT. The 3’ untranslated region of prohibitin and cellular immortalization. Exp Cell Res 1996; 224:128–35 [DOI] [PubMed] [Google Scholar]
- 36.Manjeshwar S, Branam DE, Lerner MR, Brackett DJ, Jupe ER. Tumor suppression by the prohibitin gene 3’untranslated region RNA in human breast cancer. Cancer Res 2003; 63:5251–6 [PubMed] [Google Scholar]
- 37.Yang H, Li TWH, Zhou Y, Peng H, Liu T, Zandi E, Martínez-Chantar ML, Mato JM, Lu SC. Activation of a novel c-Myc-miR27-Prohibitin 1 circuitry in cholestatic liver injury inhibits glutathione synthesis in mice. Antioxid Redox Signal 2015; 22:259–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramani K, Mavila N, Ko KS, Mato JM, Lu SC. Prohibitin 1 regulates the H19-Igf2 axis and proliferation in hepatocytes. J Biol Chem 2016; 291:24148–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajalingam K, Wunder C, Brinkmann V, Churin Y, Hekman M, Sievers C, Rapp UR, Rudel T. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration. Nat Cell Biol 2005; 7:837–43 [DOI] [PubMed] [Google Scholar]
- 40.Luan Z, He Y, Alattar M, Chen Z, He F. Targeting the prohibitin scaffold-CRAF kinase interaction in RAS-ERK-driven pancreatic ductal adenocarcinoma. Mol Cancer 2014; 13:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan TL, Wang PW. Explore the molecular mechanism of apoptosis induced by tanshinone IIA on activated rat hepatic stellate cells. Evid Based Complement Altern Med ECAM 2012; 2012:734987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma A, Qadri A. Vi polysaccharide of salmonella typhi targets the prohibitin family of molecules in intestinal epithelial cells and suppresses early inflammatory responses. Proc Natl Acad Sci USA 2004; 101:17492–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wintachai P, Wikan N, Kuadkitkan A, Jaimipuk T, Ubol S, Pulmanausahakul R, Auewarakul P, Kasinrerk W, Weng WY, Panyasrivanit M, Paemanee A, Kittisenachai S, Roytrakul S, Smith DR. Identification of prohibitin as a chikungunya virus receptor protein. J Med Virol 2012; 84:1757–70 [DOI] [PubMed] [Google Scholar]
- 44.Kuadkitkan A, Wikan N, Fongsaran C, Smith DR. Identification and characterization of prohibitin as a receptor protein mediating DENV-2 entry into insect cells. Virology 2010; 406:149–61 [DOI] [PubMed] [Google Scholar]
- 45.Liu S, Wang W, Brown LE, Qiu C, Lajkiewicz N, Zhao T, Zhou J, Porco JA, Wang TT. A novel class of small molecule compounds that inhibit hepatitis C virus infection by targeting the prohibitin-CRaf pathway. EBioMedicine 2015; 2:1600–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolonin MG, Saha PK, Chan L, Pasqualini R, Arap W. Reversal of obesity by targeted ablation of adipose tissue. Nat Med 2004; 10:625–32 [DOI] [PubMed] [Google Scholar]
- 47.Hossen MN, Kajimoto K, Akita H, Hyodo M, Ishitsuka T, Harashima H. Therapeutic assessment of cytochrome C for the prevention of obesity through endothelial cell-targeted nanoparticulate system. Mol Ther J Ther 2013; 21:533–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci USA 2002; 99:6274–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puppin C, Passon N, Franzoni A, Russo D, Damante G. Histone deacetylase inhibitors control the transcription and alternative splicing of prohibitin in thyroid tumor cells. Oncol Rep 2011; 25:393–7 [DOI] [PubMed] [Google Scholar]
- 50.Theiss AL, Obertone TS, Merlin D, Sitaraman SV. Interleukin-6 transcriptionally regulates prohibitin expression in intestinal epithelial cells. J Biol Chem 2007; 282:12804–12 [DOI] [PubMed] [Google Scholar]
- 51.Schmidt-Arras D, Rose-John S. IL-6 pathway in the liver: from physiopathology to therapy. J Hepatol 2016; 64:1403–15 [DOI] [PubMed] [Google Scholar]
- 52.Qian X, Zhao P, Li W, Shi Z-M, Wang L, Xu Q, Wang M, Liu N, Liu LZ, Jiang BH. MicroRNA-26a promotes tumor growth and angiogenesis in glioma by directly targeting prohibitin. CNS Neurosci Ther 2013; 19:804–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fletcher CE, Dart DA, Sita-Lumsden A, Cheng H, Rennie PS, Bevan CL. Androgen-regulated processing of the oncomir miR-27a, which targets prohibitin in prostate cancer. Hum Mol Genet 2012; 21:3112–27 [DOI] [PubMed] [Google Scholar]
- 54.Liu T, Tang H, Lang Y, Liu M, Li X. MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett 2009; 273:233–42 [DOI] [PubMed] [Google Scholar]
- 55.Chen W, Qi J, Bao G, Wang T, Du CW, Wang MD. Emerging role of microRNA-27a in human malignant glioma cell survival via targeting of prohibitin. Mol Med Rep 2015; 12:1515–23 [DOI] [PubMed] [Google Scholar]
- 56.Cirilo PDR, de Sousa Andrade LN, Corrêa BRS, Qiao M, Furuya TK, Chammas R, Penalva L. MicroRNA-195 acts as an anti-proliferative miRNA in human melanoma cells by targeting prohibitin 1. BMC Cancer 2017; 17:750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J, Aung LHH, Long B, Qin D, An S, Li P. miR-23a binds to p53 and enhances its association with miR-128 promoter. Sci Rep 2015; 5:16422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng F, Qiu B, Zang R, Song P, Gao S. Pseudogene PHBP1 promotes esophageal squamous cell carcinoma proliferation by increasing its cognate gene PHB expression. Oncotarget 2017; 8:29091–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han EK, Mcgonigal T, Butler C, Giranda VL, Luo Y. Characterization of Akt overexpression in MiaPaCa-2 cells: prohibitin is an Akt substrate both in vitro and in cells. Anticancer Res 2008; 28:957–63 [PubMed] [Google Scholar]
- 60.Ande SR, Gu Y, Nyomba BLG, Mishra S. Insulin induced phosphorylation of prohibitin at tyrosine 114 recruits Shp1. Biochim Biophys Acta 2009; 1793:1372–8 [DOI] [PubMed] [Google Scholar]
- 61.Ande SR, Moulik S, Mishra S. Interaction between O-GlcNAc modification and tyrosine phosphorylation of prohibitin: implication for a novel binary switch. PLoS One 2009; 4:e4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson WE, Ramalho-Santos J, Sutovsky P. Ubiquitination of prohibitin in mammalian sperm mitochondria: possible roles in the regulation of mitochondrial inheritance and sperm quality control. Biol Reprod 2003; 69:254–60 [DOI] [PubMed] [Google Scholar]
- 63.Suh SK, Hood BL, Kim BJ, Conrads TP, Veenstra TD, Song BJ. Identification of oxidized mitochondrial proteins in alcohol-exposed human hepatoma cells and mouse liver. Proteomics 2004; 4:3401–12 [DOI] [PubMed] [Google Scholar]
- 64.Ande SR, Mishra S. Prohibitin interacts with phosphatidylinositol 3,4,5-triphosphate (PIP3) and modulates insulin signaling. Biochem Biophys Res Commun 2009; 390:1023–8 [DOI] [PubMed] [Google Scholar]
- 65.Chiu CF, Ho MY, Peng JM, Hung SW, Lee WH, Liang CM, Liang SM. Raf activation by Ras and promotion of cellular metastasis require phosphorylation of prohibitin in the raft domain of the plasma membrane. Oncogene 2013; 32:777–87 [DOI] [PubMed] [Google Scholar]
- 66.Mishra S, Ande SR, Nyomba B. The role of prohibitin in cell signaling. FEBS J 2010; 277:3937–46 [DOI] [PubMed] [Google Scholar]
- 67.Santamaria E, Avila MA, Latasa MU, Rubio A, Martin-Duce A, Lu SC, Corrales FJ. Functional proteomics of nonalcoholic steatohepatitis: mitochondrial proteins as targets of S-adenosylmethionine. Proc Natl Acad Sci USA 2003; 100:3065–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev 2012; 92:1515–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murray B, Barbier-Torres L, Fan W, Mato JM, Lu SC. Methionine adenosyltransferases in liver cancer. World J Gastroenterol 2019; 25:4300–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang H, Liu T, Wang J, Li TWH, Fan W, Peng H, Krishnan A, Gores GJ, Mato JM, Lu SC. Deregulated methionine adenosyltransferase α1, c-Myc, and Maf proteins together promote cholangiocarcinoma growth in mice and humans(‡). Hepatology 2016; 64:439–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chowdhury I, Thompson WE, Thomas K. Prohibitins role in cellular survival through Ras-Raf-MEK-ERK pathway. J Cell Physiol 2014; 229:998–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang J, Li B, He QY. Significance of prohibitin domain family in tumorigenesis and its implication in cancer diagnosis and treatment. Cell Death Dis 2018; 9:580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sánchez-Quiles V, Santamaría E, Segura V, Sesma L, Prieto J, Corrales FJ. Prohibitin deficiency blocks proliferation and induces apoptosis in human hepatoma cells: molecular mechanisms and functional implications. Proteomics 2010; 10:1609–20 [DOI] [PubMed] [Google Scholar]
- 74.Monga SP. β-Catenin signaling and roles in liver homeostasis, injury, and tumorigenesis. Gastroenterology 2015; 148:1294–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nejak-Bowen KN, Monga S. Beta-catenin signaling, liver regeneration and hepatocellular cancer: sorting the good from the bad. Semin Cancer Biol 2011; 21:44–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Theiss AL, Jenkins AK, Okoro NI, Klapproth J-M, Merlin D, Sitaraman SV. Prohibitin inhibits tumor necrosis factor alpha–induced nuclear factor-kappa B nuclear translocation via the novel mechanism of decreasing importin α3 expression. Mol Biol Cell 2009; 20:4412–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luedde T, Schwabe RF. NF-κB in the liver–linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 2011; 8:108–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shimizu Y, Okada K, Tsukishiro T, Nishimori H, Higuchi K, Watanabe A. Cytokine effect on the concentrations of interleukin-8 (IL-8) in the culture medium of human liver tumor cell lines. Int Hepatol Commun 1993; 1:174–7 [Google Scholar]
- 79.Okada K, Shimizu Y, Tsukishiro T, Minemura M, Nishimori H, Higuchi K, Watanabe A. Serum interleukin-8 levels in patients with hepatocellular carcinoma. Int Hepatol Commun 1994; 2:178–82 [Google Scholar]
- 80.Akiba J, Yano H, Ogasawara S, Higaki K, Kojiro M. Expression and function of interleukin-8 in human hepatocellular carcinoma. Int J Oncol 2001; 18:257–64 [DOI] [PubMed] [Google Scholar]
- 81.Miyamoto M, Shimizu Y, Okada K, Kashii Y, Higuchi K, Watanabe A. Effect of interleukin-8 on production of tumor-associated substances and autocrine growth of human liver and pancreatic cancer cells. Cancer Immunol Immunother CII 1998; 47:47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]