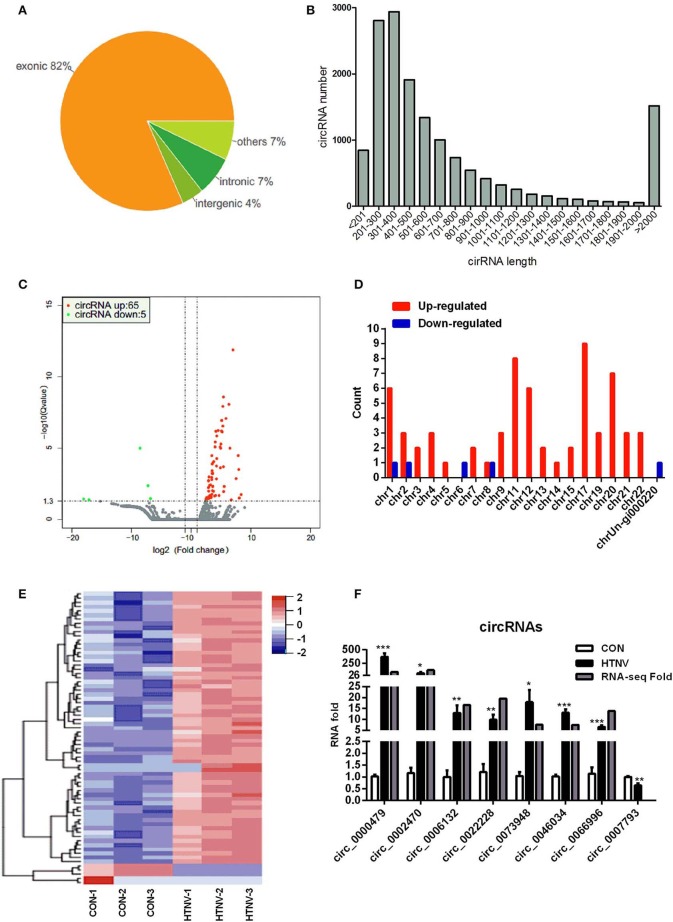
Figure 1.
circRNA expression overview. (A) circRNAs category chart. (B) circRNAs length distribution. (C) Volcano plot of DE circRNAs upon HTNV infection in HUVECs. Red dots represent up-regulated circRNAs and green dots represent down-regulated circRNAs. (D) Chromosome distribution of DE circRNAs. (E) Heatmap and clustering analysis of DE circRNAs. Each row represents one circRNA and each column represents one sample; −2, −1, 0, 1, and 2 represent fold change. Red indicates high expression and blue represents low expression. CON-1, CON-2 and CON-3 represent three mock-infected samples; HTNV-1, HTNV-2, and HTNV-3 represent three HTNV-infected samples. (F) Verification of dysregulated circRNAs. HUVECs were infected with HTNV 76-118 (MOI = 1) for 3 days. Then the total RNA was extracted and the expression levels of circRNAs were measured by RT-qPCR. Student's t-test, mean ± standard deviation (SD), *P < 0.05; **P < 0.01; ***P < 0.001. The experiment was performed at least three times independently.