Figure 1:
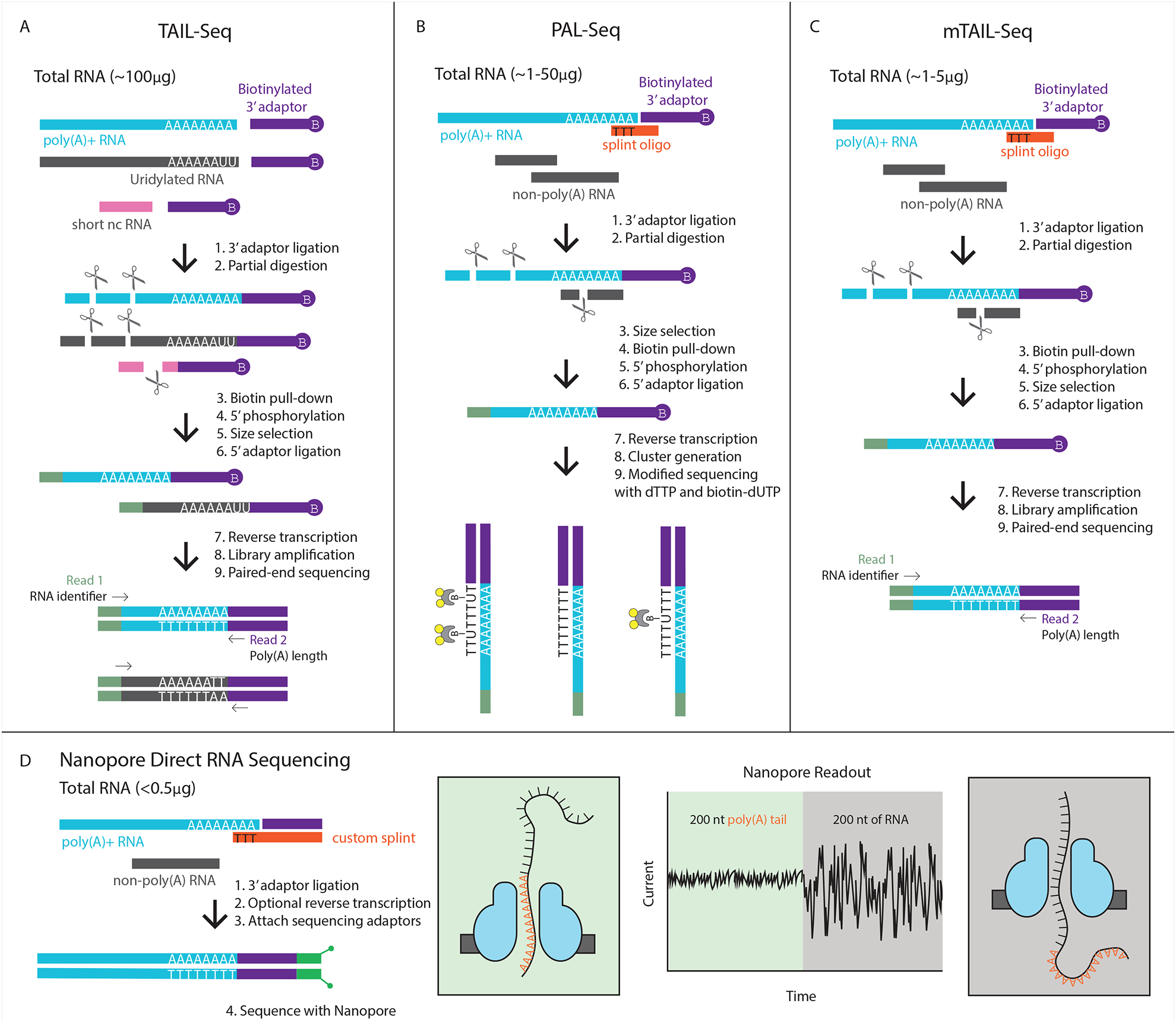
Comparison of different sequencing methods for reading poly(A) tails. A) TAIL-seq is able to capture the 3’ end of any RNA, and therefore gives a readout of both poly(A) tail length as well as other modifications such as uridylation through an innovative Tailseeker algorithm. B) PAL-seq uses a splint oligo to preferentially capture polyadenylated RNAs, thus bypassing rRNA removal. Biotin-labeled dUTP marks each cluster in proportion to the length of the tail. C) mTAIL-seq uses the splint oligo approach in order to reduce the amount of starting material needed, and uses the Tailseeker software to read poly(A) tail length. D) Nanopore technology is a new way to sequence that can be used to directly sequence RNA or cDNA with minimal library preparation needed. The nucleic acid travels through the nanopore at a constant rate; therefore, the dwell time of the poly(A) tail in the nanopore correlates to its length.
