Abstract
Background
This study aimed to investigate circular RNA‐mitochondrial tRNA translation optimization 1 (circ‐MTO1) expression in tumor tissue and its correlation with clinical characteristics and survival profiles, as well as its effect on cancer cell functions in prostate cancer.
Methods
A total of 298 primary prostate cancer patients were included. Reverse transcription‐quantitative polymerase chain reaction was conducted to evaluate circ‐MTO1 expression in tumor tissue and paired adjacent tissue. Disease‐free survival (DFS) and overall survival (OS) were recorded. In in vitro experiment, prostate cancer cells were transfected with circ‐MTO1 over‐expression and negative‐control over‐expression plasmids. Then cell proliferation, cell invasion and miR‐630 as well as miR‐17‐5p expressions in prostate cancer cells were detected.
Results
Circular RNA‐mitochondrial tRNA translation optimization 1 expression was downregulated in tumor tissue compared with paired adjacent tissue (P < .001) in patients with prostate cancer. Circ‐MTO1 high expression in tumor tissue was correlated with decreased pathological T stage (P = .001) as well as lower pathological N stage (P = .020). As for survival profiles, the DFS (P = .006) and OS (P = .018) were both longer in patients who had circ‐MTO1 high expression compared with patients who had circ‐MTO1 low expression. In addition, circ‐MTO1 high expression independently predicted favorable DFS and OS. Besides, further in vitro experiments illustrated that circ‐MTO1 inhibited proliferation (P < .05) and invasion (P < .05) as well as downregulated miR‐17‐5p expression in prostate cancer cells (P < .05).
Conclusion
Circ‐MTO1 correlates with decreased pathological T/N stage and favorable survival profiles, and it also inhibits cell proliferation, invasion as well as miR‐17‐5p expression in prostate cancer.
Keywords: cell function, circ‐MTO1, clinical characteristics, prostate cancer, survival profile
1. INTRODUCTION
Prostate cancer, ranked as the second most frequent cancer in men worldwide, attacks roughly 1.3 million people and leads to 359 000 deaths in 2018 according to the most recent global cancer statistical report.1 Diagnosis of prostate cancer mainly bases on biopsy, and although new imaging technology has been progressed to enhance the diagnostic accuracy, effective screening and timely diagnosis are still insufficient.2, 3, 4 Treatment of prostate cancer has sufficiently progressed, while, for the early stage patients presenting with good response to treatment and favorable survival, they often suffer from sequelae or adverse events. As for the patients in late stage with unsatisfactory response to treatment, their survival profile is still poor.5, 6, 7 Therefore, it is necessary to explore more biomarkers, which could assist with the management of prostate cancer.
Circular RNAs (circRNAs), a novel class of non‐coding RNAs presenting with closed loops without poly A tails, are stably expressed in human and have almost no protein‐coding function.8 In recent years, circRNAs have been revealed as genetic factors that play critical roles in various cancers, such as promoting cancer progression by regulating cancer cell functions and presenting with potential as prognostic biomarkers.9, 10, 11 Additionally, circRNAs are increasingly implicated in prostate cancer as well; however, the relevant studies are not as abundant as the studies of other cancers.12, 13 Circular RNA‐mitochondrial tRNA translation optimization 1 (circ‐MTO1) is a novel circRNA that is previously reported as an anti‐tumor gene, which inhibits liver fibrosis to prevent hepatocellular carcinoma and represses progression of lung cancer and breast cancer.14, 15 And circ‐MTO1 has also been reported to express in prostate cancer (http://www.circbase.org/), Based on these facts, we supposed that circ‐MTO1 might play a crucial role in prostate cancer.
Thus, the aim of this study was to investigate circ‐MTO1 expression in tumor tissue and its correlation with clinical characteristics, survival profiles, as well as its effect on cancer cell functions in prostate cancer.
2. MATERIALS AND METHODS
2.1. Patients
A total of 298 primary prostate cancer patients who underwent surgical resection in our hospital were consecutively enrolled from January 2015 to December 2018. The inclusion criteria were: (a) histologically diagnosed as primary prostate cancer; (b) about to receive surgical resection; (c) agreed with the collection of their tumor tissue and paired adjacent tissue for the study using; (d) age ≥18 years old; (e) can be followed up regularly. Besides, the patients complicated with other malignancies were excluded. This study was approved by the Ethics Committee of our hospital and conducted according to the Declaration of Helsinki. All patients signed the informed consents before enrollment.
2.2. Data and sample collection
The major tumor characteristics of patients were recorded including Gleason score, pathological T stage, pathological N stage, surgical margin status and prostate‐specific antigen (PSA) level. The fresh tumor tissue and paired adjacent tissue excised from surgery were divided into two parts. One was sent to the pathology department for routine pathologic analysis, and the other was snap‐frozen and used for circ‐MTO1 (access number: has_circ_0076979) determination in this study. And the relative expression of circ‐MTO1 in tumor tissue and adjacent tissue was detected by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR).
2.3. Treatment and follow‐up
After enrollment, preoperative treatments were administered to patients as appropriate based on the patients' clinical status, then surgery was carried out. After surgery, conventional care and adjuvant therapy were given to patients as well, and both the preoperative treatment and the adjuvant therapy were conducted according to the guideline,16 which were not intervened by the current study. Regular follow‐up was performed by telephone or clinic visit. The last follow‐up date was 2018/12/31, and the median follow‐up duration was 26.5 months (range: 0.0‐48.0 months). Disease‐free survival (DFS) was calculated from the date of entry into the study to the date of relapse or death. For the patients not known to have relapsed or died at last follow‐up, they were censored on the date of last examination. Overall survival (OS) was calculated from the date of entry into the study to the date of death. For patients not known to have died at last follow‐up, they were censored on the date of last known to be alive.
2.4. Cell culture
Human prostate cancer cell lines including DU‐145, VCaP and PC‐3 as well as human normal prostate epithelial cell line RWPE‐1 were purchased from Cell Bank of Type Culture Collection of Chinese Academy of Sciences. DU‐145 cells were cultured in 90% minimum eagle's medium (MEM) (Gibco) and 10% fetal bovine serum (FBS) (Gibco), VCaP cells were cultured in 90% Dulbecco's modified eagle medium (DMEM) (Gibco) and 10% FBS (Gibco), PC‐3 cells were cultured in 90% Ham's F‐12 Nutrient Mix medium (F‐12) and 10% FBS (Gibco), and RWPE‐1 cells were cultured in Keratinocyte Serum Free Medium (K‐SFM) Kit (Invitrogen, USA). All cells were maintained in a humidified tissue culture incubator at 37°C in 5% CO2. After culturing, the relative expression of Circ‐MTO1 in Human prostate cancer cell lines were detected by RT‐qPCR, with the RWPE‐1 cells used as control.
2.5. Transfection and assays
The circ‐MTO1 over‐expression plasmids and negative‐control over‐expression plasmids were respectively transfected into the PC‐3 cells, which were constructed by Shanghai Qeejen Bio‐Tech Co., Ltd using pEX2. And the PC‐3 cells transfected with circ‐MTO1 over‐expression plasmids were named as OE‐Circ group, and the PC‐3 cells transfected with negative‐control over‐expression plasmids were named as OE‐NC group, correspondingly. At 24 hours (h) after transfection, the circ‐MTO1 relative expression in two groups were detected by RT‐qPCR, meanwhile, the cell invasive ability in two groups was evaluated by the transwell assay; at 0 hour, 24 hours, 48 hours and 72 hours after transfection, cell proliferation in two groups was determined by CCK‐8 (Dojindo) assay, which was carried out in accordance with the manufacturer's instructions and assessed by optical density (OD) value. In addition, previous studies demonstrated that microRNA‐630 (miR‐630) and microRNA‐17‐5p (miR‐17‐5p) were directly targeted by the circ‐MTO1.14 Consequently, the relative expressions of miR‐17‐5p and miR‐630 in two groups were also detected at 24 hours after transfection by the RT‐qPCR.
2.6. RT‐qPCR
In the beginning, the RNA extraction from tissues and cells was carried out by RNeasy Protect Mini Kit (Qiagen) according to the instructions of the manufacturer. Then for circ‐MTO1 detection, the linear RNA was digested using RNase R (Epicentre), while for miR‐17‐5p and miR‐630 detection the linear RNA was not digested. Subsequently, the RNA was reversely transcribed into cDNA by the iScript cDNA Synthesis Kit (Bio‐Rad), afterward, PCR was performed by QuantiNova SYBR Green PCR Kit (Qiagen). Then, the relative expressions of circ‐MTO1 and miR‐17‐5p/miR‐630 were calculated using the formula 2−ΔΔCt. Besides, the internal reference for circ‐MTO1 was GAPDH, and the internal reference for miR‐17‐5p as well as miR‐630 was U6. And before the detection of GAPDH, the linear RNA was digested by RNase R (Epicentre). In addition, all the primers used in RT‐qPCR were as follows: Circ‐MTO1 Forward: TTACCAGCCGAGTAGAGTTCC; Reverse: ATCCATTCCTTCAGGTTCCAAC; miR‐17‐5p Forward: ACACTCCAGCTGGGTGAGGTAGTAGGTTGTGT; Reverse: TGTCGTGGAGTCGGCAATTC; miR‐630 Forward: ACACTCCAGCTGGGAGTATTCTGTACCAGG; Reverse: TGTCGTGGAGTCGGCAATTC; GAPDH Forward: GGAGCGAGATCCCTCCAAAAT; Reverse: GGCTGTTGTCATACTTCTCATGG; U6 Forward: CTCGCTTCGGCAGCACATATACTA; Reverse: ACGAATTTGCGTGTCATCCTTGC.
2.7. Statistical analysis
Statistical analysis was performed using SPSS 24.0 (IBM), and figures were plotted using GraphPad Prism 7.00 (GraphPad Software). Categorical variables were summarized using count and percentage, while continuous variables were expressed as mean and standard deviation (SD) or median and interquartile range (IQR). Comparison between paired samples was determined by Wilcoxon signed‐rank test; Comparison between independent samples was determined by one‐way analysis of variance (ANOVA) followed by Dunnett's t test, unpaired t test, Wilcoxon rank‐sum test, or chi‐square test, as appropriate. DFS and OS were displayed by Kaplan‐Meier curves, and the difference of DFS and OS between groups was determined by log‐rank test. Variables predicting DFS and OS were analyzed by univariate and multivariate Cox's proportional hazard regression model. P value <.05 was considered as significant.
3. RESULTS
3.1. Baseline characteristics
The mean age was 62.0 ± 9.5 years in patients with prostate cancer enrolled in our study (Table 1). And the numbers of patients with Gleason score ≤6, =7 and ≥8 were 63 (21.2%), 167 (56.0%) as well as 68 (22.8%), respectively. In addition, the number of patients with pathological T stage of pT2, pT3, and pT4 were 176 (59.1%), 116 (38.9%), and 6 (2.0%), respectively, and there were 216 (72.5%) patients with pathological N stage of pN0 as well as 82 (27.5%) patients with pN1. As for the surgical margin status, there were 241 (80.9%) patients who had a negative margin and 57 (19.1%) patients who had a positive status. The number of patients who had a PSA level ≤10, 10‐20 and ≥20 were 78 (26.2%), 161 (54.0%) and 59 (19.8%), respectively.
Table 1.
Clinical characteristics of prostate cancer patients
| Items | Prostate cancer patients (N = 298) |
|---|---|
| Age (y), mean ± SD | 62.0 ± 9.5 |
| Gleason score, No. (%) | |
| ≤6 | 63 (21.2) |
| =7 | 167 (56.0) |
| ≥8 | 68 (22.8) |
| Pathological T stage, No. (%) | |
| pT2 | 176 (59.1) |
| pT3 | 116 (38.9) |
| pT4 | 6 (2.0) |
| Pathological N stage, No. (%) | |
| pN0 | 216 (72.5) |
| pN1 | 82 (27.5) |
| Surgical margin status, No. (%) | |
| Negative | 241 (80.9) |
| Positive | 57 (19.1) |
| PSA (ng/mL), No. (%) | |
| ≤10 | 78 (26.2) |
| 10‐20 | 161 (54.0) |
| ≥20 | 59 (19.8) |
Abbreviations: PSA, prostate‐specific antigen; SD, standard deviation.
3.2. Expression of circ‐MTO1 in tumor tissue and paired adjacent tissue
The expression of circ‐MTO1 in tumor tissue and paired adjacent tissue was detected by RT‐qPCR, which displayed that circ‐MTO1 expression was lower in tumor tissue compared with paired adjacent tissue in prostate cancer patients (P < .001) (Figure 1).
Figure 1.
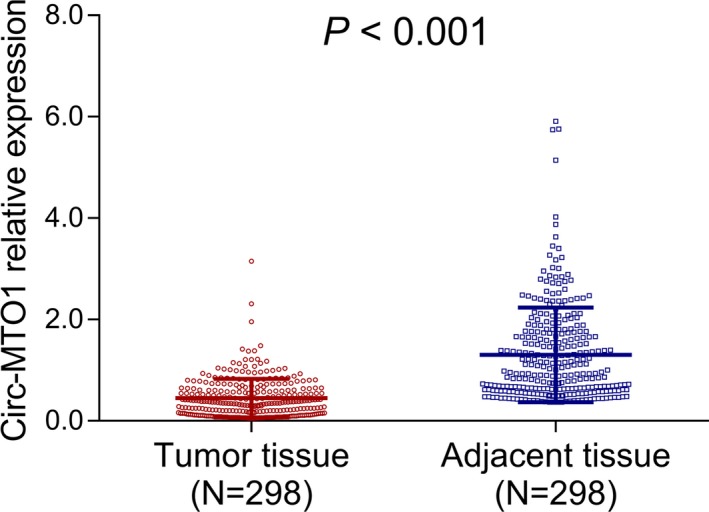
Circ‐MTO1 expression in tumor tissue and paired adjacent tissue. The comparison of circ‐MTO1 expression between tumor tissue and paired adjacent tissue obtained from prostate cancer patients. Comparison was determined by Wilcoxon signed‐rank test. P value <.05 was considered as significant. Circ‐MTO1, circular RNA mitochondrial tRNA translation optimization 1
3.3. Correlation of circ‐MTO1 expression in tumor tissue with clinical features
The circ‐MTO1 high expression in tumor tissue was correlated with decreased pathological T stage (P = .001) and lower pathological N stage (P = .020) in patients with prostate cancer, while it was not associated with age (P = .345), Gleason score (P = .793), surgical margin status (P = .185) or PSA level (P = .806) (Table 2).
Table 2.
Correlation of circ‐MTO1 expression with clinical characteristics
| Items | Circ‐MTO1 expressiona | P value | |
|---|---|---|---|
| Low | High | ||
| Age (y), No. (%) | |||
| <60 | 56 (46.7) | 64 (53.3) | .345 |
| ≥60 | 93 (52.2) | 85 (47.8) | |
| Gleason score, No. (%) | |||
| ≤6 | 31 (49.2) | 32 (50.8) | .793 |
| =7 | 83 (49.7) | 84 (50.3) | |
| ≥8 | 35 (51.5) | 33 (48.5) | |
| Pathological T stage, No. (%) | |||
| pT2 | 74 (42.0) | 102 (58.0) | .001 |
| pT3 | 70 (60.3) | 46 (39.7) | |
| pT4 | 5 (83.3) | 1 (16.7) | |
| Pathological N stage, No. (%) | |||
| pN0 | 99 (45.8) | 117 (54.2) | .020 |
| pN1 | 50 (61.0) | 32 (39.0) | |
| Surgical margin status, No. (%) | |||
| Negative | 116 (48.1) | 125 (51.9) | .185 |
| Positive | 33 (57.9) | 24 (42.1) | |
| PSA (ng/mL), No. (%) | |||
| ≤10 | 39 (50.0) | 39 (50.0) | .806 |
| 10‐20 | 82 (50.9) | 79 (49.1) | |
| ≥20 | 28 (47.5) | 31 (52.5) | |
Correlation was determined by Wilcoxon rank‐sum test or chi‐square test.
Abbreviation: PSA, prostate‐specific antigen.
High and low were classified by the median value of circ‐MTO1 relative expression in tumor tissues.
3.4. Correlation of tumor tissue circ‐MTO1 with survival profiles
The DFS was more favorable in patients who had circ‐MTO1 high expression in tumor tissue compared with prostate cancer patients who had circ‐MTO1 low expression (P = .006) (Figure 2A), and the OS was also longer in patients with circ‐MTO1 high expression than that in patients with circ‐MTO1 low expression (P = .018) (Figure 2B). Furthermore, univariate and multivariate Cox's regression analyses were performed to evaluate the factors predicting DFS. Univariate Cox's regression revealed that circ‐MTO1 high expression (P = .005) predicted increased DFS, while higher Gleason score (P = .027), higher pathological T stage (P = .001), higher pathological N stage (P = .002) as well as positive surgical margin (P = .004) predicted worse DFS (Table 3). Then, all the factors were included in the multivariate Cox's regression analysis, which disclosed that circ‐MTO1 high expression (P = .032) could independently predict increased DFS. However, higher Gleason score (P = .017), higher pathological T stage (P = .005), and positive surgical margin (P = .012) were independent factors for predicting shorter DFS. In terms of OS, univariate Cox's regression analysis illuminated that circ‐MTO1 high expression (P = .016) predicted better OS, while higher Gleason score (P = .001), higher pathological N stage (P < .001), positive surgical margin (P = .003) and higher PSA level (P = .029) predicted worse OS (Table 4). Subsequently, all the factors were analyzed by multivariate Cox's regression analysis, which illustrated that circ‐MTO1 high expression (P = .044) independently predicted more prolonged OS, while higher Gleason score (P < .001), higher pathological N stage (P = .028), positive surgical margin (P = .020), and higher PSA level (P = .013) were independent predictive factors for less satisfactory OS.
Figure 2.
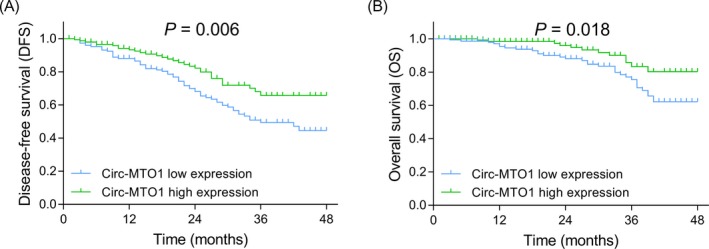
DFS and OS in patients with circ‐MTO1 low or high expressions. The comparison of DFS between patients with circ‐MTO1 low expression and patients with circ‐MTO1 high expression (A), the comparison of OS between patients with circ‐MTO1 low expression and patients with circ‐MTO1 high expression (B). DFS and OS were displayed by Kaplan‐Meier curves, and the difference of DFS and OS between the two groups was determined by log‐rank test. P value <.05 was considered as significant. DFS, disease‐free survival; OS, overall survival; Circ‐MTO1, circular RNA mitochondrial tRNA translation optimization 1
Table 3.
Analysis of factors predicting DFS
| Items | Univariate Cox's regression | Multivariate Cox's regression | ||
|---|---|---|---|---|
| P value | HR (95% CI) | P value | HR (95% CI) | |
| Circ‐MTO1 high expression | .005 | 0.541 (0.351‐0.832) | .032 | 0.611 (0.389‐0.959) |
| Age ≥60 y | .305 | 1.247 (0.818‐1.902) | .327 | 1.244 (0.804‐1.923) |
| Higher Gleason score | .027 | 1.408 (1.039‐1.909) | .017 | 1.454 (1.069‐1.976) |
| Higher pathological T stage | .001 | 1.847 (1.278‐2.670) | .005 | 1.730 (1.177‐2.541) |
| Higher pathological N stage | .002 | 1.937 (1.279‐2.934) | .106 | 1.447 (0.924‐2.267) |
| Positive surgical margin | .004 | 1.932 (1.227‐3.040) | .012 | 1.817 (1.139‐2.898) |
| Higher PSA level | .211 | 1.205 (0.899‐1.615) | .103 | 1.278 (0.952‐1.715) |
Factors predicting DFS was analyses by univariate and multivariate Cox's proportional hazard regression model.
Abbreviations: CI, confidence interval; DFS, disease‐free survival; HR, hazard ratio; PSA, prostate‐specific antigen.
Table 4.
Analysis of factors predicting OS
| Items | Univariate Cox's regression | Multivariate Cox's regression | ||
|---|---|---|---|---|
| P value | HR (95% CI) | P value | HR (95% CI) | |
| Circ‐MTO1 high expression | .016 | 0.444 (0.229‐0.860) | .044 | 0.490 (0.244‐0.982) |
| Age ≥60 y | .212 | 1.484 (0.798‐2.758) | .465 | 1.280 (0.661‐2.479) |
| Higher Gleason score | .001 | 2.049 (1.317‐3.188) | <.001 | 2.298 (1.456‐3.626) |
| Higher pathological T stage | .055 | 1.670 (0.988‐2.821) | .090 | 1.629 (0.926‐2.865) |
| Higher pathological N stage | <.001 | 2.976 (1.653‐5.358) | .028 | 2.127 (1.087‐4.164) |
| Positive surgical margin | .003 | 2.504 (1.360‐4.612) | .020 | 2.170 (1.129‐4.170) |
| Higher PSA level | .029 | 1.605 (1.051‐2.451) | .013 | 1.740 (1.124‐2.693) |
Factors predicting OS was analyzed by univariate and multivariate Cox's proportional hazard regression model.
Abbreviations: CI, confidence interval; HR, hazard ratio; OS, overall survival; PSA, prostate‐specific antigen.
3.5. Effect of circ‐MTO1 on prostate cancer cell proliferation and invasion as well as its potential target miRNA
The circ‐MTO1 expression was downregulated in DU‐145 cells (P < .05), VCaP cells (P < .01) and PC‐3 cells (P < .001) compared with RWPE‐1 cells (Figure 3A). Then PC‐3 cells were selected for further cellular experiments. After transfections, the circ‐MTO1 expression was elevated in OE‐Circ group compared with OE‐NC group (P < .001) (Figure 3B). In view of cell proliferation, the cell proliferation rate of PC‐3 cells was decreased in OE‐Circ group compared with OE‐NC group at 48 hours (P < .05) and 72 hours (P < .05) after transfections (Figure 3C). And the invasive cell count was reduced in OE‐Circ group compared with OE‐NC group at 24 hours after transfections (P < .01) (Figure 3D,E). For the reason that miR‐17‐5p and miR‐630 were reported as direct targets of circ‐MTO1 by previous studies, we subsequently detected the expressions of miR‐17‐5p and miR‐630 in OE‐Circ group and OE‐NC group, and the results displayed that the miR‐17‐5p (P < .01) (Figure 3F) expression in OE‐Circ group was decreased than that in OE‐NC group, while the expression of miR‐630 (P > .05) (Figure 3G) was of no difference between the two groups.
Figure 3.
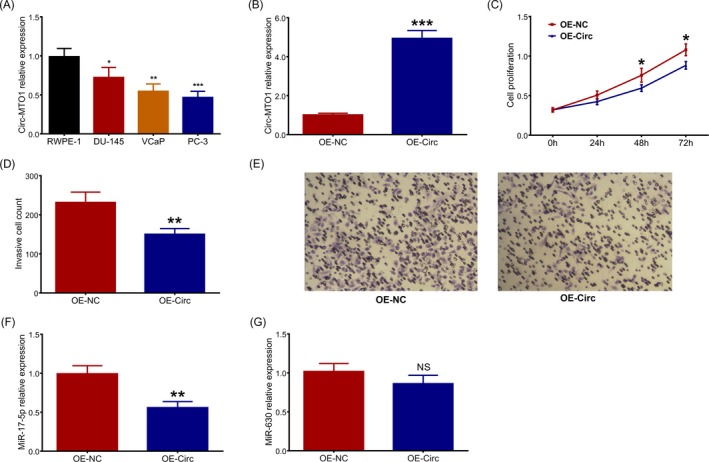
Effect of circ‐MTO1 on proliferation, invasion and miR‐17‐5p/miR‐630 in prostate cancer cells. Circ‐MTO1 expression in prostate cancer cells lines and human normal prostate epithelial cell line (A), circ‐MTO1 expression in EC‐Circ group and OE‐NC group after transfections in PC‐3 cells (B), effect of circ‐MTO1 on cell proliferation in PC‐3 cells (C), effect of circ‐MTO1 on cell invasive count in PC‐3 cells (D, E), effect of circ‐MTO1 on miR‐17‐5p (F) and miR‐630 (G) expressions in PC‐3 cells. Comparison between the two groups was determined by Dunnett's t test. P value <.05 was considered as significant. Circ‐MTO1, circular RNA‐mitochondrial tRNA translation optimization 1; ANOVA, one‐way analysis of variance
4. DISCUSSION
In this study, we discovered that (a) circ‐MTO1 expression was downregulated in tumor tissue compared with non‐tumor tissue in prostate cancer; (b) circ‐MTO1 high expression in tumor tissue correlated with less severe clinical characteristics in patients with prostate cancer; (c) circ‐MTO1 high expression in tumor tissue associated with more prolonged DFS and OS and was an independent predicting factor for favorable DFS and OS; and (d) circ‐MTO1 repressed cell proliferation and invasion as well as downregulated miR‐17‐5p expression in prostate cancer cells.
Evidence correlating circRNAs with the pathogenesis of prostate cancer is preliminary but promising. For instance, circ‐ABCC4 advocates the progression of prostate cancer by acting as competing endogenous RNA (ceRNA) of miR‐1182 via promoting the expression of forkhead box P4.12 Another study reveals that circ_0001206 is decreased in prostate cancer tumor tissue and represses cell proliferation, migration and invasion in prostate cancer cells.17 Additionally, a recent research discloses that the decreased circ‐itchy E3 ubiquitin‐protein ligase associates with more severe pathological T stage, increased lymph node metastasis risk and worse survival in patients with prostate cancer.18 In terms of the specific circRNA (circ‐MTO1) evaluated in our study, the circ‐MTO1, it has not been studied in prostate cancer before. However, there have been several studies demonstrate the anti‐oncogenetic role of circ‐MTO1 in other cancers. For example, a previous in vitro study reports that circ‐MTO1/miR‐17/QKI‐5 regulatory circuit suppresses lung cancer cell proliferation.19 Furthermore, circ‐MTO1 inhibits cancer cell viability and reverses the resistance to monastrol in breast cancer cells.15 In this study, we found that circ‐MTO1 was downregulated in tumor tissue compared with non‐tumor tissue, which might be resulted from that circ‐MTO1 decreased malignant cell proliferation as shown in our in vitro experiments. Thus, tumor tissue presented with a lower circ‐MTO1 expression compared with the non‐tumor tissue as the tumor cell was featured by its malignant cell proliferation ability. And we also discovered that circ‐MTO1 high expression in tumor tissue was correlated with decreased pathological T stage and N stage, which could be explained by that circ‐MTO1 might act as a tumor suppressor in prostate cancer via suppressing cancer cell proliferation and invasion as displayed in our following in vitro experiment. In addition, we also discovered that circ‐MTO1 high expression associated with better DFS and OS and was an independent predictive factor for favorable DFS and OS. Here are several possible explanations: a) circ‐MTO1 possibly delayed prostate cancer progression by reducing cancer cell proliferation which led to attenuated tumor growth, or inhibiting cancer cell invasion which resulted in less severe tumor metastasis, via the interaction with other factors, for instance, the interaction with miR‐17 and QKI‐5. In addition, we also observed that circ‐MTO1 suppressed prostate cancer cell proliferation, invasion and downregulated miR‐17‐5p expression in prostate cancer cells, which might also provide some explanation to these results. b) Circ‐MTO1 might also inhibit prostate cancer progression by repressing chemoresistance, which contributed to a better treatment response and subsequent favorable survival.15, 19
Increasing information about the effect of circRNAs on cancer cell functions in prostate cancer has been revealed recently. For example, circ‐SMARCA5 is an androgen‐responsive circRNA that is overexpressed and enhances cell proliferation in prostate cancer cells.20 And circ‐nyosin light chain kinase enhances prostate cancer cell proliferation, invasion and migration by mediating the level of miR‐29a.21 Although the detailed mechanism of circ‐MTO1 in mediating cell activities in prostate cancer has not been investigated in our study, we observed that circ‐MTO1 inhibited cell proliferation and invasion, and also downregulated miR‐17‐5p expression in prostate cancer cells. MiR‐17‐5p has been reported to be correlated with cancer development and progression. For instance, miR‐17 enhances cell proliferation and migration of human colorectal cancer cells via decreasing salt‐inducible kinase 1.22 Moreover, downregulation of miR‐17 reduces chemoresistance and represses EMT in a death effector domain‐containing DNA‐binding protein‐dependent manner in gastric cancer cells.23 MiR‐17‐5p is also reported to promote cancer cell proliferation via changing cell cycle profiles by dysregulating the retinoblastoma‐like protein 2/E2F4‐repressing complexes.24 More importantly, miR‐17‐5p has also been reported as an oncological gene in prostate cancer as well. A previous study elucidates that miR‐17‐5p promotes prostate cancer cell proliferation, colony formation, cell survival, and cell invasion by targeting TIMP metallopeptidase inhibitor 3.14 These previous studies indicate the role of miR‐17 in promoting disease progression in various carcinomas, including prostate cancer. Thus, we speculated that circ‐MTO1 repressed cell proliferation and invasion possibly by downregulating miR‐17‐5p. However, this speculation still needs to be validated by further mechanistic experiments.
Furthermore, there were several limitations in this study: (a) the follow‐up duration was relatively short, which should be prolonged in the future; we only assessed the expression of circ‐MTO1 in tumor tissue but not in circulating samples that were more convenient considering the clinical utility, such as plasma or serum, which should be evaluated in the future study; (b) patients enrolled in this study were prostate cancer patients who had underwent resection; therefore, the role of circ‐MTO1 in unresectable prostate cancer patients was unclear.
In conclusion, circ‐MTO1 correlates with decreased pathological T/N stage and favorable survival profiles, and it also inhibits cell proliferation, invasion as well as miR‐17‐5p expression in prostate cancer.
ACKNOWLEDGMENTS
None.
Hu Y, Guo B. Circ‐MTO1 correlates with favorable prognosis and inhibits cell proliferation, invasion as well as miR‐17‐5p expression in prostate cancer. J Clin Lab Anal. 2020;34:e23086 10.1002/jcla.23086
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 3. Hayes JH, Barry MJ. Screening for prostate cancer with the prostate‐specific antigen test: a review of current evidence. JAMA. 2014;311(11):1143‐1149. [DOI] [PubMed] [Google Scholar]
- 4. Eastham J. Prostate cancer screening. Investig Clin Urol. 2017;58(4):217‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hamdy FC, Donovan JL, Lane JA, et al. 10‐year outcomes after monitoring, surgery, or radiotherapy for localized prostate cancer. N Engl J Med. 2016;375(15):1415‐1424. [DOI] [PubMed] [Google Scholar]
- 6. Skolarus TA, Wolf AM, Erb NL, et al. American Cancer Society prostate cancer survivorship care guidelines. CA Cancer J Clin. 2014;64(4):225‐249. [DOI] [PubMed] [Google Scholar]
- 7. Litwin MS, Tan HJ. The diagnosis and treatment of prostate cancer: a review. JAMA. 2017;317(24):2532‐2542. [DOI] [PubMed] [Google Scholar]
- 8. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381‐388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tanaka E, Miyakawa Y, Kishikawa T, et al. Expression of circular RNA CDR1AS in colon cancer cells increases cell surface PDL1 protein levels. Oncol Rep. 2019;42(4):1459‐1466. [DOI] [PubMed] [Google Scholar]
- 10. Verduci L, Strano S, Yarden Y, et al. The circRNA‐microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13(4):669‐680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pan Y, Xu T, Liu Y, Li W, Zhang W. Upregulated circular RNA circ_0025033 promotes papillary thyroid cancer cell proliferation and invasion via sponging miR‐1231 and miR‐1304. Biochem Biophys Res Commun. 2019;510(2):334‐338. [DOI] [PubMed] [Google Scholar]
- 12. Huang C, Deng H, Wang Y, et al. Circular RNA circABCC4 as the ceRNA of miR‐1182 facilitates prostate cancer progression by promoting FOXP4 expression. J Cell Mol Med. 2019;23(9):6112‐6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen D, Lu X, Yang F, Xing N. Circular RNA circHIPK3 promotes cell proliferation and invasion of prostate cancer by sponging miR‐193a‐3p and regulating MCL1 expression. Cancer Manag Res. 2019;11:1415‐1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang W, Dong R, Guo Y, et al. CircMTO1 inhibits liver fibrosis via regulation of miR‐17‐5p and Smad7. J Cell Mol Med. 2019;23(8):5486‐5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Y, Dong Y, Zhao L, Su L, Luo J. Circular RNAMTO1 suppresses breast cancer cell viability and reverses monastrol resistance through regulating the TRAF4/Eg5 axis. Int J Oncol. 2018;53(4):1752‐1762. [DOI] [PubMed] [Google Scholar]
- 16. NCCN clinical practice guidelines in Oncology: prostate Cancer. https://www.nccn.org/
- 17. Song Z, Zhuo Z, Ma Z, Hou C, Chen G, Xu G. Hsa_Circ_0001206 is downregulated and inhibits cell proliferation, migration and invasion in prostate cancer. Artif Cells Nanomed Biotechnol. 2019;47(1):2449‐2464. [DOI] [PubMed] [Google Scholar]
- 18. Huang E, Chen X, Yuan Y. Downregulated circular RNA itchy E3 ubiquitin protein ligase correlates with advanced pathologic T stage, high lymph node metastasis risk and poor survivals in prostate cancer patients. Cancer Biomark. 2019;26(1):41‐50. [DOI] [PubMed] [Google Scholar]
- 19. Zhang B, Chen M, Jiang N, Shi K, Qian R. A regulatory circuit of circ‐MTO1/miR‐17/QKI‐5 inhibits the proliferation of lung adenocarcinoma. Cancer Biol Ther. 2019;20(8):1127‐1135. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Kong Z, Wan X, Zhang Y, et al. Androgen‐responsive circular RNA circSMARCA5 is up‐regulated and promotes cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;493(3):1217‐1223. [DOI] [PubMed] [Google Scholar]
- 21. Dai Y, Li D, Chen X, et al. Circular RNA Myosin Light Chain Kinase (MYLK) promotes prostate cancer progression through modulating Mir‐29a expression. Med Sci Monit. 2018;24:3462‐3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang C, Liu J, Xu L, et al. MicroRNA‐17 promotes cell proliferation and migration in human colorectal cancer by downregulating SIK1. Cancer Manag Res. 2019;11:3521‐3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu DM, Hong XW, Wang LL, et al. MicroRNA‐17 inhibition overcomes chemoresistance and suppresses epithelial‐mesenchymal transition through a DEDD‐dependent mechanism in gastric cancer. Int J Biochem Cell Biol. 2018;102:59‐70. [DOI] [PubMed] [Google Scholar]
- 24. Zhu Y, Gu J, Li Y, et al. MiR‐17‐5p enhances pancreatic cancer proliferation by altering cell cycle profiles via disruption of RBL2/E2F4‐repressing complexes. Cancer Lett. 2018;412:59‐68. [DOI] [PubMed] [Google Scholar]


