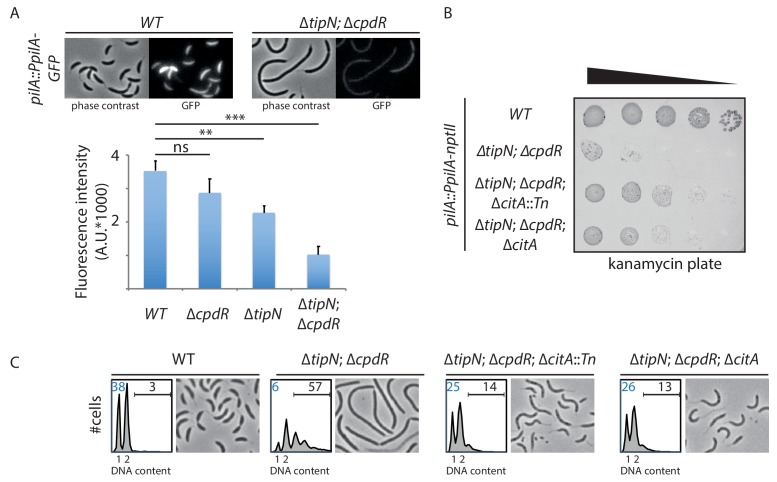
Figure 2. Genetic screen to identify Tn insertions that enhance CtrA.
(A) CtrA activity in WT (MB2325), ∆tipN (MB2337) and ∆cpdR (MB2329) single mutant cells, and in ∆tipN ∆cpdR (MB2331) double mutant cells, was monitored using a pilA::PpilA-GFP transcriptional reporter whose activity is dependent on CtrA. Fluorescence intensity was automatically quantified, and t-tests were performed to determine the significance with p<0.05 (**) and p<0.005 (***). (B) Spot dilutions of the indicated strains (MB2268 [WT], MB2271 [∆tipN; ∆cpdR], MB3056 [∆tipN; ∆cpdR; ∆citA::Tn], and MB3058 [∆tipN; ∆cpdR; ∆citA] from top to bottom) carrying the pilA::PpilA-nptII transcriptional reporter on PYE plates containing kanamycin (20 µg.mL−1). (C) FACS profiles and phase contrast images of the strains shown in panel (B). FACS profiles showing genome content (ploidy) of cells growing exponentially in PYE and then treated with rifampicin (20 µg.mL−1) for 3 hours to inhibit DNA replication. Numbers (%) of G1-phase cells and cells containing more than two chromosomes are indicated in blue and black, respectively.
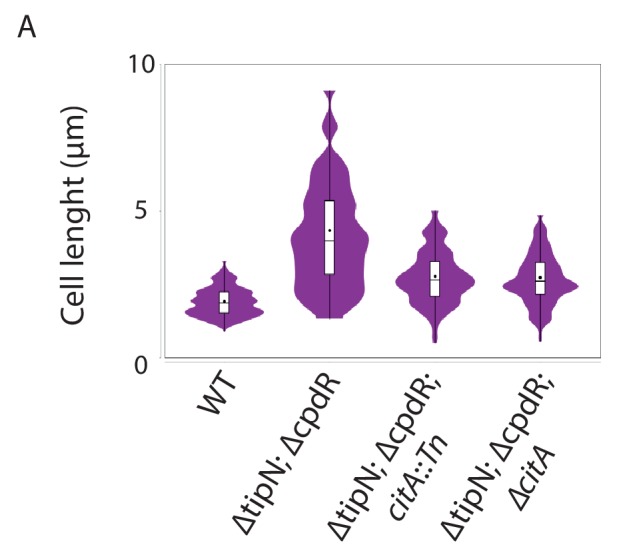
Figure 2—figure supplement 1. Loss of CitA attenuates the cell division defect of tipN cpdR double mutant cells.