Matrix-bound nanovesicles (MBVs) are a novel subclass of extracellular vesicles.
Abstract
Biomaterials composed of extracellular matrix (ECM) provide both mechanical support and a reservoir of constructive signaling molecules that promote functional tissue repair. Recently, matrix-bound nanovesicles (MBVs) have been reported as an integral component of ECM bioscaffolds. Although liquid-phase extracellular vesicles (EVs) have been the subject of intense investigation, their similarity to MBV is limited to size and shape. Liquid chromatography–mass spectrometry (LC-MS)–based lipidomics and redox lipidomics were used to conduct a detailed comparison of liquid-phase EV and MBV phospholipids. Combined with comprehensive RNA sequencing and bioinformatic analysis of the intravesicular cargo, we show that MBVs are a distinct and unique subpopulation of EV and a distinguishing feature of ECM-based biomaterials. The results begin to identify the differential biologic activities mediated by EV that are secreted by tissue-resident cells and deposited within the ECM.
INTRODUCTION
The development of extracellular matrix (ECM)–based biomaterials as surgical meshes, topical powders, and injectable hydrogels is a relatively recent pursuit compared to similar efforts with synthetic biomaterials (1). The evolutionarily conserved composition of the ECM, and its critical role in development, cell, tissue, and organ homeostasis, and the response to tissue injury provide a compelling rationale for its use as an inductive biomaterial to promote the repair of damaged tissues and organs (2). However, gaps in our understanding of the dynamic biophysical properties and composition of ECM limit our ability to design and fabricate ECM-based biomaterials that fully capitalize upon their therapeutic potential. Although advanced proteomic techniques have begun to provide an accurate profile of ECM fibrillar and soluble protein components (2), the effects of constitutive (phospho)lipids and incorporated (extracellular) RNA on matrix biology and the host tissue response following injury are largely unknown.
Extracellular vesicles (EVs) are nanometer-sized vesicles encapsulated by a membrane, which transport cell signaling molecules including microRNA (miRNA), (phospho)lipids, and proteins (3). As a potent mediator of cell signaling, EVs have been the subject of intense investigation and thought to be secreted exclusively into a liquid phase where they can be relocated freely between cells and to distant sites using biologic fluids as a mobile medium. Recently, the presence of EV firmly embedded within the ECM, termed matrix-bound nanovesicles (MBVs), was described (4). In contrast to liquid-phase EV, MBVs are secreted by tissue-resident cells and integrated into the fibrillar matrix, a feature that may define their biological purpose and mechanism of action. The nature and biologic significance of the MBV cargo and lipid membrane has not been characterized. Given that MBVs are integrated within the matrix, it is plausible that the molecular speciation of their constituent phospholipids may facilitate this interaction.
The objective of the present study is to identify similarities and differences between liquid-phase (i.e., exosomes) and matrix-bound forms (i.e., MBV) of EV. However, given that EV present in biological fluids, MBV present in native tissue ECM, and ECM-based biomaterials represent heterologous populations secreted from multiple cell sources, a direct comparative in vivo analysis between these putative EV populations is problematic. As an alternative to using body fluid or tissue-derived vesicles, ECM and conditioned medium produced in vitro by cultured cells can be isolated (5). This approach offers several advantages such as the use of a single cell type source, thereby obviating any doubts regarding vesicle origin, the ability to selectively harvest vesicles from either liquid- or solid-phase compartments, and the ability to control the cell culture environment and thus also control vesicle composition and cargo.
Here, we use a fibroblast cell culture model that allows selective harvesting of liquid-phase EV and MBV integrated into the matrix. We conduct RNA sequencing and bioinformatic analysis to characterize the differential miRNA signature and use liquid chromatography–mass spectrometry (LC-MS)–based lipidomics and redox lipidomics protocols to perform detailed analysis of molecular speciation of liquid-phase EV and MBV phospholipids. Results from these comparative material analyses show that MBVs represent a subpopulation of nanovesicle distinct from EV found in a liquid phase and significantly expand our understanding of EV and matrix biology. Separately, the use of ECM-based therapies can now be examined by new perspectives, and MBV can assist in the design of next-generation ECM-based materials. For example, given their nanometer size, MBV can be used in minimally invasive applications that are otherwise untenable for decellularized ECM scaffolds from which MBVs are derived, such as intravitreal injections to prevent ischemia-induced retinal ganglion cell axon degeneration (6), or incorporated into existing ECM-based devices for sustainable release of vesicles (7, 8). Furthermore, results of the present study may aid in the future development of EV-based theranostic biomaterials such as artificial EV for clinical use (9).
RESULTS
Isolation of liquid-phase EV and MBV
We used scanning electron microscopy (SEM) to provide high-resolution, high-magnification imaging of MBV embedded within an ECM bioscaffold derived from porcine urinary bladder matrix (UBM). SEM images revealed discrete spheres approximately 100 nm in diameter dispersed throughout the collagen fibers (Fig. 1A). Given their compartmentalization within ECM scaffolds, we hypothesized that MBVs deposited into a solid ECM substrate are a unique class of EV separate from EV secreted into a liquid phase. To test this hypothesis, we used an in vitro 3T3 fibroblast cell culture model that allows selective harvesting of vesicles from a liquid-phase or solid-phase extracellular compartment (Fig. 1B). Representative images from phase-contrast microscopy, and hematoxylin and eosin (H&E)– and 4′,6-diamidino-2-phenylindole–stained sections showed that no residual cells or intact nuclei were visible after decellularization of the cell culture plate (Fig. 1C). Transmission electron microscopy (TEM) imaging of liquid-phase EV harvested from the cell culture supernatant (Fig. 1D) and MBV isolated from decellularized ECM (Fig. 1E) showed that these two populations of vesicles shared a similar morphology. Moreover, nanoparticle tracking analysis (NTA) distribution plots showed similar vesicle size of both liquid-phase EV and MBV, with the majority of vesicles having a diameter of <200 nm (Fig. 1F). To determine whether MBV contained markers commonly attributed to exosomes, immunoblot analysis was performed for CD63, CD81, CD9, and Hsp70 (10). Results showed that, in contrast to liquid-phase EV, the MBV showed a marked decrease in CD63, CD81, and CD9 (Fig. 1G). Furthermore, silver staining of electrophoretically separated proteins showed that MBV contained a protein cargo that was distinctly different than the liquid-phase EV (Fig. 1H), suggesting that MBV may be a unique subpopulation of nanovesicle.
Fig. 1. Isolation of liquid-phase EV and MBV.
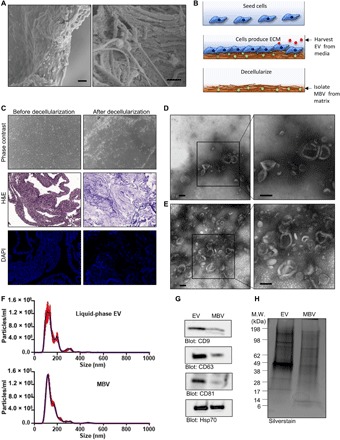
(A) SEM images of an ECM scaffold derived from urinary bladder matrix (UBM) showing discrete spherical bodies approximately 100 nm in diameter dispersed throughout the matrix. Scale bars, 1 μm. (B) Illustration of the 3T3 fibroblast cell culture model used to selectively harvest vesicles from a liquid-phase or solid-phase extracellular compartment. (C) Phase-contrast microscopy, hematoxylin and eosin (H&E) staining, and 4′,6-diamidino-2-phenylindole (DAPI) staining showing the absence of cells and intact cell nuclei after decellularization. (D and E) TEM of liquid-phase EV (D) and MBV (E) isolated from the 3T3 fibroblast cell culture model. Scale bars, 100 nm. (F) Size distribution plots from nanoparticle tracking analysis (NTA) of liquid-phase EV (top) and MBV (bottom) isolates from the 3T3 fibroblast cell culture. (G) Immunoblot analysis of CD9, CD63, CD81, and Hsp70 expression levels in liquid-phase EV and MBV. (H) Silverstain analysis of electrophoretically separated proteins in liquid-phase EV and MBV. M.W., molecular weight.
miRNA is selectively packaged into liquid-phase EV and MBV derived from 3T3 fibroblasts
We used comprehensive next-generation RNA-sequencing (RNA-seq) to catalog differentially expressed miRNA in MBV and liquid-phase EV relative to the 3T3 fibroblast parent cell from which these vesicles were derived. Bioanalyzer analysis revealed the absence of 18S and 28S ribosomal RNA, and an enrichment of small RNA molecules [<200 nucleotides (nt)] in total RNA isolated from liquid-phase EV and MBV. However, the small RNA size distribution from liquid-phase EV was much broader than MBV with a marked enrichment of small RNA molecules between 100 and 200 nt in liquid-phase EV (Fig. 2A). We focused the analysis on differential miRNA signatures by conducting next-generation sequencing of miRNA libraries generated from the parental cellular RNA, the liquid-phase EV, and the MBV isolates (n = 3 per group). Principal components analysis (PCA) showed that within respective groups, the replicate miRNA profiles clustered close to one another (Fig. 2B). However, extensive differences in miRNA content were observed between the parental cell and the liquid-phase EV and MBV isolates. Overall, 28 (50.91%) miRNAs were found to be differentially expressed in MBV compared to liquid-phase EV by at least twofold (Fig. 2C). In addition, respective liquid-phase EV or MBV and the parental cellular miRNA profiles were clearly distinct (Fig. 2, B and C). To validate the results of miRNA sequencing, reverse transcription quantitative polymerase chain reaction (RT-qPCR) was conducted to detect three up-regulated miRNAs (miR-163-5p, miR-27a-5p, and miR-92a-1-5p) and three down-regulated miRNAs (miR-451a, miR-93b-5p, miR-99b-5p) in MBV compared to liquid-phase EV isolated from 3T3 fibroblasts (Fig. 2D). As anticipated, the results showed that the level of miR-163-5p, miR-27a-5p, and miR-92a-1-5p was up-regulated, and miR-451a, miR-93b-5p, and miR-99b-5p was down-regulated in MBV compared to liquid-phase EV, thereby corroborating the results from the miRNA sequencing data. Ingenuity Pathway Analysis (IPA) of differentially enriched miRNAs in MBV compared to liquid-phase EV showed a strong association with organ and system development and function. In contrast, miRNAs differentially enriched in liquid-phase EV compared to MBV were associated with pathways involved in cellular growth, development, proliferation, and morphology (Fig. 2E).
Fig. 2. miRNA is selectively packaged into liquid-phase EV and MBV.
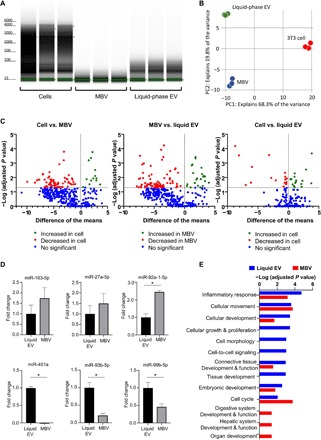
(A) Bioanalyzer analysis of total RNA isolated from 3T3 parental cells and their secreted liquid-phase EV and MBV. (B) Principal components analysis (PCA) comparing liquid-phase EV (green), MBV (blue), and cellular (red) RNA-seq datasets. (C) Volcano plot showing the differential expression of miRNAs in liquid-phase EV, MBV, and the parental cells. The inclusion criteria were a twofold difference of log2 (fold change) in either direction with a P value of <0.05. Each dot represents a specific miRNA transcript; green dots to the right of the vertical dashed line correspond to a relative increase in expression level, and red dots to the left correspond to a relative decrease in expression level. Blue dots indicate miRNA with no significant change in expression level. (D) RT-qPCR validation of the results of miRNA sequencing. *P < 0.05, n = 4. (E) IPA functional analysis. Significantly enriched molecular functions identified by IPA functional analysis considering differentially expressed miRNA in MBV (red) and liquid-phase EV (blue).
MBV miRNA content is unique to the cellular origin
Results with the 3T3 fibroblast cell model showed selective packaging of miRNA within MBV deposited in the ECM compared to liquid-phase EV secreted into the cell culture supernatant. We next sought to determine whether MBV miRNA cargo is unique to the cellular origin. We characterized and compared the miRNA composition of MBV isolated from ECM produced in vitro by bone marrow–derived stem cells (BMSCs), adipose stem cells (ASCs), and umbilical cord stem cells (UCSCs) isolated from different human donors using next-generation sequencing methods. A representative phase-contrast microscopy image of a decellularized BMSC cell culture plate showed the absence of cells and the presence of branched fibrillar structures (Fig. 3A). TEM imaging of isolated MBV from a decellularized BMSC cell culture plate showed the characteristic morphology attributed to EV (Fig. 3B). Furthermore, NTA showed similar distribution plots between BMSC-, ASC-, and UCSC-derived MBV, with the majority of vesicles having a diameter of <200 nm (Fig. 3, C to E). After isolation of total RNA from these samples, bioanalyzer analysis showed the absence of ribosomal RNA and an enrichment of small RNA molecules (<200 nt) (Fig. 3F). miRNA libraries were generated from the samples (BMSC, n = 3 human donors; ASC, n = 3 human donors; UCSC, n = 3 human donors) and subjected to miRNA sequencing. A PCA showed that samples clustered primarily by the cell type from which they were derived (Fig. 3G). Of note, despite the use of three separate human donors for each cell type used to generate the MBV samples, the PCA showed a high degree of homogeneity in the miRNA profile within the respective groups (Fig. 3G). In addition, volcano plots showed that fewer miRNAs were found differentially expressed between BMSC- and UCSC-derived MBV than between BMSC-ASC and UCSC-ASC.
Fig. 3. MBV miRNA content is unique to the cellular origin.
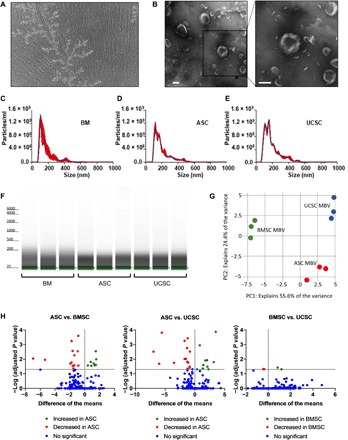
(A) Phase-contrast microscopy image of a decellularized BMSC cell culture plate showing the absence of cells. (B) TEM of MBV isolated from the decellularized BMSC culture plate. Scale bars, 100 nm. (C to E) Size distribution plots from NTA of MBV isolated from BMSC (C), ASC (D), and UCSC (D) decellularized culture plates. (F) Bioanalyzer analysis of total RNA isolated from BMSC-, ASC-, and UCSC-derived MBV. (G) PCA comparing BMSC MBV (green), UCSC MBV (blue), and ASC MBV (red) RNA-seq datasets. (H) Volcano plot showing the differential expression of miRNAs in BMSC-, ASC-, and UCSC-derived MBV. The inclusion criterion was a twofold difference of log2 (fold change) in either direction with a P value of <0.05. Each dot represents a specific miRNA transcript; green dots to the right of the vertical dashed line correspond to a relative increase in expression level, and red dots to the left correspond to a relative decrease in expression level. Blue dots indicate miRNA with no significant change in expression level.
Phospholipid profiles of liquid-phase EV, MBV, and parent cells
Several studies have characterized the lipid composition of EV (11). However, there are no data on phospholipid composition of MBV. Therefore, we performed LC-MS–based global lipidomics and redox lipidomics analyses to comparatively evaluate the phospholipid composition of MBV and liquid-phase EV compared to their 3T3 fibroblast parent cells (Fig. 4, A and D). Nine major phospholipid classes were detected in all three types of samples, with the total number of detected molecular species of 536 distributed between the following major classes: bis-monoacylglycerophosphate (BMP), 59 species; phosphatidylglycerol (PG), 37 species; cardiolipin (CL), 117 species; phosphatidylinositol (PI), 33 species; phosphatidylethanolamine (PE), 102 species; phosphatidylserine (PS), 45 species; phosphatidic acid (PA), 26 species; phosphatidylcholine (PC), 107 species; and sphingomyelin (SM), 10 species (Fig. 4D). In terms of their content of polyunsaturated fatty acid (PUFA) residues, PE, PI, PC, and PS represented the major reservoir of these polyunsaturated phospholipid species containing four to seven double bonds (Fig. 4B). These PUFA phospholipids represent the likely precursors of the signaling lipid mediators. The formation of the mediators occurs via the catalytic oxygenation of PUFA phospholipids by 5-lipoxygenase or 15-lipoxygenase to yield oxygenated phospholipids that are subsequently hydrolyzed by one of specialized phospholipases A2 to release oxygenated fatty acids, i.e., lipid mediators (12, 13). In addition, oxidized PUFA phospholipids act as signaling molecules coordinating many intracellular processes and cell responses, including apoptosis, ferroptosis, and inflammation (14). We found significant differences in molecular speciation of these phospholipids and their relative contents between liquid-phase EV and MBV (Fig. 4E). With a notable exception of SM, arachidonic acid (AA) and docosahexaenoic acid (DHA) residues were detected in all phospholipids (Fig. 4E). For many of the phospholipids, the amounts were significantly higher in MBV versus liquid-phase EV and parent cells (Fig. 4E), which identify MBV as a rich reservoir of PUFA phospholipids. PUFA phospholipids can be hydrolyzed by phospholipase A2 (PLA2), resulting in the release of free PUFA and lysophospholipids (LPLs) (15). The former can be further used by two major oxygenases, cyclooxygenase (COX) and lipoxygenase (LOX), to produce lipid mediators with pro- or anti-inflammatory capacities (13, 16, 17). This finding qualifies MBV as potential precursors for synthesis of these lipid mediators dependently on the cell/tissue context (13). Quantitatively, MBVs were enriched in PI, PS, PG, and BMP (Fig. 4C and table S1). In contrast, the content of PE, PA, and SM was higher in liquid-phase EV. PC was a predominant phospholipid in cells and liquid-phase EV. The content of a unique mitochondrial phospholipid, CL, was significantly lower in liquid-phase EV compared to MBV and parent cells (Fig. 4F). Because CL is a unique mitochondria-specific phospholipid localized predominantly in the inner mitochondrial membrane (18), this finding represents a possible link of the MBV biogenesis with the mitochondrial compartment of cells. Plasmalogen phospholipids (or ether phospholipids) are structurally different from diacyl phospholipids (or ester phospholipids) (19). In plasmalogens, vinyl ether bond is linking the sn-1 saturated or monounsaturated chain to the glycerol backbone of phospholipids (19). It has been shown that ether lipids, PE, and PC plasmalogens can facilitate membrane fusion (20) and increase membrane thickness of EV (21, 22) and therefore may play a role in nanovesicle uptake by cells. Detailed MS/MS analysis showed a high level of ether PE and PC species (plasmalogens) in both liquid-phase EV and MBV. These species were identified as PE-16:0p/20:4, PE-16:1p/20:4, PE-18:1p/20:4, PE-18:1p/22:6 and PC-16:0p/20:4, PC-18:0p/20:4, PC-20:0p/20:4, PC-18:0p/22:6, respectively (Fig. 4E).
Fig. 4. LC-MS characterization of phospholipids demonstrates their molecular speciation and differences between MBV, liquid-phase EV, and the parent cells.
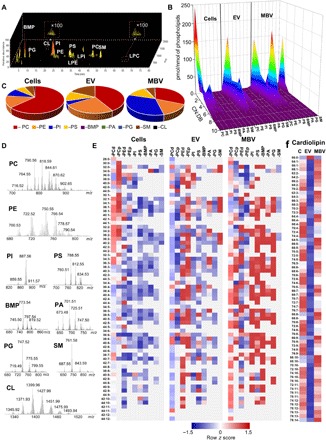
(A) Typical total ion chromatogram of phospholipids obtained from MBV. (B) Mass spectra of the major phospholipid classes in MBV. Quantitative assessment of saturated (double bond number = 0), monounsaturated (double bond number = 1), and polyunsaturated (double bond number = 2 to 10) species of phospholipids. (C) Pie plots showing the total content of major phospholipids. Data are presented as percentage of total phospholipids. (D to F) Contents of different phospholipid molecular species. Data are presented as heat maps, autoscaled to z scores, and coded blue (low values) to red (high values). EV, exosomal vesicles; MBV, matrix-bound vesicles; PC, phosphatidylcholine; PCd, PC diacyl species; PCp, PC plasmalogens; PE, phosphatidylethanolamine; PEd, PE diacyl species; PEp, PE plasmalogens; PI, phosphatidylinositol; PS, phosphatidylserine; BMP, bis-monoacylglycerophosphate; PA, phosphatidic acid; PG, phosphatidylglycerol; SM, sphingomyelin.
LPL profiles of liquid-phase EV, MBV, and parent cells
LPLs, hydrolytic metabolites of phospholipids created by phospholipases A, are bioactive signaling molecules that modulate a variety of physiological responses, including macrophage activation (23), inflammation and fibrosis (24), tissue repair and remodeling (25), and wound healing (26). LC-MS analysis showed that LPLs were present in all three types of samples, albeit with their total content in MBV and liquid-phase EV being 1.7 to 1.8 times greater compared to the parent cells. More specifically, seven classes of LPL have been identified: lysophosphatidylethanolamine (LPE), lysophosphatidylcholine (LPC), lysophosphatidylserine (LPS), lysophosphoinositol (LPI), lysophosphatidic acid (LPA), lysophosphatidylglycerol (LPG), and monolysocardiolipin (mCL) (Fig. 5A). MBVs were enriched in LPE, LPA, and LPG compared to parent cells (Fig. 5B). The content of LPI and mCL was significantly lower in MBV and liquid-phase EV versus cells. We found that the contents of LPA and LPG were significantly higher in MBV compared to EV. The levels of mCL and LPI in MBV were 3 and 6.3 times higher than in EV but 3.3 and 1.9 times lower compared to cells (Fig. 5, C and D). No significant changes in the contents of LPE, LPC, and LPS between MBV and EV were found. The non-oxidizable molecular species containing 16:0, 16:1, 18:0, and 18:1 were the major types found in all LPL species detected (Fig. 5C). As LPL may act as fusogenic lipids facilitating the transfer of the vesicular contents into cellular targets, this important role of LPL found in MBV should be further explored.
Fig. 5. LC-MS characterization reveals that MBVs are enriched in LPE, LPA, and LPG compared to parent cells.
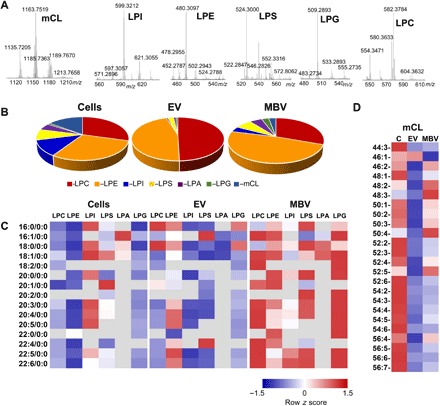
(A) Typical mass spectra of major LPL obtained from MBV. (B) Pie plots showing the total content of major LPL. Data are presented as percentage of total LPL. (C and D) Contents of LPL molecular species. Data are presented as heat maps, autoscaled to z scores, and coded blue (low values) to red (high values). N = 3. LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; LPI, lysophosphatidylinositol; LPS, lysophosphatidylserine; LPA, lysophosphatidic acid; LPG, lysophosphatidylglycerol; mCL, monolysocardiolipin.
Free fatty acids of MBV and liquid-phase EV
Given that exposure of murine bone marrow–derived macrophages to MBV results in expression of M2-like markers, Fizz1 and Arg1, which are associated with a constructive macrophage phenotype (4), we performed LC-MS analysis of PUFA and their oxygenated products in MBV versus liquid-phase EV and parent cells. MBVs were strongly enriched in AA (20:4), DHA (22:6), and docosapentaenoic fatty acids (DPA; 22:5) (Fig. 6A). In other words, MBVs represent a reservoir of substrates for the biosynthesis of signaling lipid mediators by the respective enzymatic mechanisms, COXs and LOXs. In liquid-phase EV, the major PUFAs were linoleic (18:2) and linolenic (18:3) acids (Fig. 6A).
Fig. 6. The levels of PUFA-containing phospholipids and their oxidatively modified molecular species are higher in MBV compared to those in liquid-phase EV.
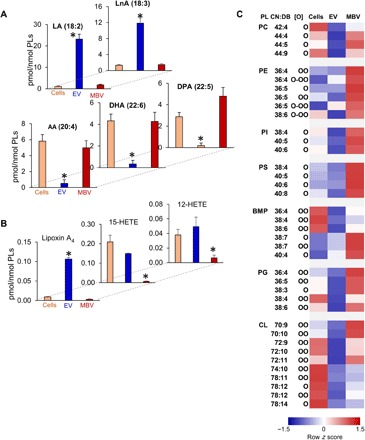
(A and B) Content of free PUFA (A) and their oxygenated metabolites (B) in parent cell, liquid-phase EV, and MBV. Data are means ± SD. *P < 0.05 versus cells or MBV. N = 3. (C) Contents of singly, doubly, and triply oxygenated phospholipid species in parent cells, liquid-phase EV, and MBV. Data are presented as heat maps, autoscaled to z scores, and coded blue (low values) to red (high values). PL, phospholipids; CL, cardiolipin.
Oxygenated fatty acids and phospholipids in MBV and liquid-phase EV
As EVs contain enzymatic machinery for biosynthesis of AA-derived lipid mediators (27), redox lipidomics analysis of oxygenated fatty acids was performed. Higher levels of AA metabolites such as 12-HETE (12-hydroxy-eicosatetraenoic acid), 15-HETE (15-hydroxy-eicosatetraenoic acid), and lipoxin A4 (LXA4) were found in liquid-phase EV versus MBV (Fig. 6B). In the context of tissue repair, LXA4 and D-series resolvin D1 (RvD1), produced by 12/15-LOX from AA (20:4) and DHA (22:6), stimulate macrophage activation to the M2-like phenotype (28). Last, we characterized oxidized phospholipids containing oxygenated AA and DHA in MBV and liquid-phase EV. The levels of oxygenated species were higher in MBV than in liquid-phase EV, where PS, PI, and PC were represented by mono-oxygenated species. BMP, PG, and CL contained singly and doubly oxygenated AA and DHA residues; triply oxygenated PUFAs were found only in PE (Fig. 6C). Overall, lipidomics and oxidative lipidomics results show that the levels of free AA, DHA, and DPA and PUFA-containing phospholipids as well as their oxidatively modified molecular species are higher in MBV than those in liquid-phase EV.
DISCUSSION
To date, research regarding EV has been largely focused on their presence in biological fluids and their potential as biomarkers of disease, with lesser emphasis upon their therapeutic potential. These liquid-phase EVs are broadly categorized based on their biogenesis, morphology, density, or cargo (29). Although it has been shown that liquid-phase EVs contain surface proteins that can mediate binding with cells and ECM molecules (30), the integration of MBV within the fibrillar network of the ECM and trafficking of MBV across all cell membranes is largely unexplored. Traditional in vitro cell culture models neglect to differentiate the liquid-phase EV secreted into the cell culture supernatant from the MBV embedded within the ECM. Here, we fractionated vesicle populations based on their compartmentalization into either the liquid-phase cell culture medium or the solid-phase ECM substrate. In terms of composition, we found that MBV isolated from the ECM of 3T3 fibroblasts contained a differential miRNA and lipid signature compared with liquid-phase EV and with the parent cell. These data are suggestive of a scenario in which molecular sorting occurs during vesicle biogenesis to specifically distribute miRNA and lipids to vesicles destined for different extracellular locations. Moreover, the cell’s capacity to differentiate between a liquid interface and a solid substrate and to selectively deposit tailored subpopulations of vesicles with distinct lipid signatures into these disparate compartments provides evidence for a different and independent membrane biogenesis of MBV from the biogenesis of EV secreted into a liquid phase. Considering that MBVs were shown to be integrated within the dense fibrillar network of the ECM, it is plausible that MBVs are secreted by cells in concert with ECM components during matrix deposition, tissue development and homeostasis, and dynamic matrix remodeling following injury. Furthermore, given that the ECM is a complex mixture of proteins, proteoglycans, and glycosaminoglycans arranged in a tissue-specific three-dimensional architecture (2), it is logical that MBV cargo and lipid content are also unique to the tissue and cellular origin. We have previously shown that MBVs isolated from ECM bioscaffolds derived from anatomically distinct source tissue have differential miRNA signatures (4). Results from the present study further support this hypothesis in that MBV isolated from ECM produced in vitro by BMSC, ASC, and UCSC derived from different human donors contained a distinctive miRNA signature specific to the cell source. In addition, fewer miRNAs were found differentially expressed between BMSC- and UCSC-derived MBV than between BMSC-ASC and UCSC-ASC, a finding that may be attributed to tissue-specific differentiation potentials of ASC (31). These findings further underline the cell-specific features of MBV miRNA profiles, which were not significantly affected by the intrinsic variability of donors. However, given that the three human donors were all male, further studies to determine sex-related variations in the miRNA cargo of MBV from the stem cell samples are warranted. Gender-specific differential expression of exosomal miRNA has been observed in human subjects (32). Gender variation is just one aspect for further investigation since other variables including age (33) and disease state (34) have been shown to affect EV miRNA cargo. PCA showed a high degree of batch-to-batch consistency of the miRNA cargo from MBV deposited by specific cell types isolated from different human donors, a finding that has important implications for MBV and ECM biomaterial manufacturing for use as research tools or clinical therapeutics. For example, a major challenge in the production of EV or ECM biomaterials for preclinical or clinical use is the standardization of product characterization to meet regulatory requirements for batch consistency and for reproducibility of the manufacturing process (35, 36). Stated differently, the observed batch-to-batch consistency in miRNA cargo from MBV deposited by specific cell types may lend itself to the development of markers to verify identity and purity of MBV intended for clinical testing. Although the present study establishes that MBV integrated into the matrix are a unique subpopulation of EV, we cannot rule out the possibility of heterogeneity within the MBV subset. A similar heterogeneity has recently been described for EV secreted into cell culture medium by mesenchymal stem cells, which were shown to secrete at least three types of liquid-phase EV that could be differentially isolated based on their affinities for membrane lipid–binding ligands (37). In addition, although MBV showed a marked decrease in proteins commonly attributed to exosomes (e.g., CD63, CD81, and CD9), future proteomics studies will be required to identify the differential expression of surface and luminal proteins associated with liquid-phase EV compared to MBV.
As the composition and compartmentalization are different for liquid-phase EV and MBV integrated into the matrix, these subpopulations of vesicles are likely to have different biological functions. IPA network analysis of differentially enriched miRNAs in MBV compared to liquid-phase EV isolated from the 3T3 fibroblast model showed that miRNAs in MBV are associated with significant network-associated functions in organ and system development compared to miRNA enriched from liquid-phase EV. MBVs have previously been shown to recapitulate functional and phenotypical properties attributed to the ECM bioscaffolds from which they are derived, including stem cell differentiation and activation of an anti-inflammatory and pro-resolving macrophage phenotype, both of which are hallmarks of constructive tissue remodeling (2).
In contrast to EV that are secreted into body fluids and readily available for cell-cell communication, MBVs embedded within tissue ECM are stably associated with the matrix and can only be isolated following degradation of the ECM material (4). The requirement for matrix degradation to release MBV may partially define their mechanism of action, including those related to their capacity to generate pro-resolving lipid mediators. Because MBVs remain intact and attached to ECM even after decellularization, the molecular speciation of their constituent phospholipids likely plays a role in facilitating such MBV-ECM interactions. Using LC-MS–based lipidomics and redox lipidomics approaches, we performed detailed characterization of the molecular speciation of MBV phospholipids, LPL, and the oxygenated and non-oxygenated PUFA and can speculate upon the relationship of these various molecular species with their utilization as metabolic lipid signaling platforms. We report that high levels of LPL, bioactive molecules that are important for macrophage differentiation, tissue repair, remodeling, and wound healing, are a characteristic feature of MBV. In addition, as fusogenic lipids, LPL can facilitate the transfer of the vesicular contents to intracellular targets. MBVs, but not liquid-phase EVs, were enriched in PUFA non-oxygenated and oxygenated phospholipids and therefore represent a potential reservoir of oxidized and oxidizable esterified phospholipid species, the role of which has not yet been elucidated. Notably, PUFA-enriched MBV can be viewed as an important source of lipid mediators activated by different phospholipases dependent on the pro-/anti-inflammatory context of the extracellular environment.
A limitation of the present study is the use of a single cell line to evaluate differences in liquid-phase EV and MBV cargo. The 3T3 fibroblast cell line used in this study was chosen because it is a well-characterized and widely used cell line in biologic research. Furthermore, results from the RNA-seq and lipidomic analyses showed that multiple replicates of MBV derived from the 3T3 fibroblast model showed a high level of consistency in terms of miRNA and lipid cargo and that this cargo is significantly different from the cargo of the corresponding liquid-phase EV, which supports the fidelity of our results. However, further studies are required with other cell types, including primary cells, before one can derive a more definitive understanding of the biologic purpose of MBV versus liquid-phase EV. In addition, additional studies are warranted to identify and determine the biologic relevance of MBV in native (nondecellularized) tissues.
The findings of the present study may have significant clinical implications. EVs harvested from biological fluids have been used for diagnostic purposes (38), and EVs isolated from cell culture supernatant are being explored as therapeutic agents in early-phase clinical trials (39, 40). However, delineation of the MBV subpopulation may now allow new perspectives on EV-based therapeutics, especially in the design and manufacture of novel biomaterials and artificial EV for clinical use. Given the selective loading of specific miRNA and lipid cargo within MBV, further studies on MBV biogenesis may prove to be instrumental in guiding new strategies for cargo loading of EV (9) or for the incorporation of EV into ECM-based biomaterials to be used as an inductive substrate for tissue repair (7, 8).
MATERIALS AND METHODS
Experimental design
The objective of the present study was to conduct a comparative material analysis of EV secreted into a liquid medium versus MBV integrated into the ECM using an in vitro 3T3 fibroblast cell culture model that allows selective harvesting of vesicles from liquid-phase or solid-phase extracellular compartments. 3T3 fibroblasts were seeded on polystyrene plates in the presence of ascorbic acid to induce deposition of ECM (41, 42). After 7 days, the cell culture medium containing liquid-phase EV was harvested, and the culture plates were decellularized to remove cells while maintaining the molecular composition and ultrastructure of the ECM. Following decellularization, MBVs were isolated from decellularized ECM by enzymatic digestion. We used LC-MS–based lipidomics and redox lipidomics to perform detailed analysis of liquid-phase EV and MBV phospholipids and conducted comprehensive RNA-seq and bioinformatic analysis of the intravesicular miRNA cargo.
Preparation of in vitro cell-derived ECM
Human BMSC, human ASC, and human UCSC ECM plates were provided by StemBioSys (San Antonio, TX) and prepared according to a published protocol (43). Briefly, human BMSCs, human ASCs, or human UCSCs were seeded onto a 75-cm2 cell culture flask coated with human fibronectin (1 hour at 37°C) at a cell density of 3500 cells/cm2 and cultured in α-minimum essential medium (α-MEM) supplemented with 20% fetal bovine serum (FBS) and 1% penicillin-streptomycin for 14 days. The medium was refreshed the day after initial seeding and then every 3 days. At day 7, ascorbic acid 2-phosphate (Sigma-Aldrich) was added to the medium at a final concentration of 50 μM. At day 14, plates were decellularized using 0.5% Triton in 20 mM ammonium hydroxide for 5 min and rinsed two times with Hanks’ balanced salt solution containing both calcium and magnesium (HBSS +/+) and once with ultrapure H2O. Murine NIH 3T3 fibroblast cells were seeded onto a 75-cm2 cell culture flask at a cell density of 3500 cells/cm2 and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with exosome-depleted FBS (44), 1% penicillin-streptomycin, and ascorbic acid 2-phosphate (Sigma-Aldrich) at a final concentration of 50 μM for 7 days. At day 7, the supernatant from cultured 3T3 fibroblast cells was collected, and the culture plates were washed three times with phosphate-buffered saline (PBS), decellularized using 0.5% Triton in 20 mM ammonium hydroxide for 5 min, and then rinsed three times with ultrapure H2O.
Isolation of MBV and liquid-phase EV
MBVs were isolated as previously described with minor modifications (4) . Briefly, the decellularized ECM was enzymatically digested with Liberase DL (100 ng/ml; Roche) in buffer [50 mM tris (pH 7.5), 5 mM CaCl2, and 150 mM NaCl] for 1 hour at 37°C. The cell culture supernatant containing the liquid-phase EV and the digested ECM containing the MBV were subjected to differential centrifugation at 500g (10 min), 2500g (20 min), and 10,000g (30 min), and the supernatant was passed through a 0.22-μm filter (Millipore). The clarified supernatant containing the liberated MBV or liquid-phase EV was then centrifuged at 100,000g (Beckman Coulter Optima L-90K Ultracentrifuge) at 4°C for 70 min to pellet the vesicles. The vesicle pellets were then washed and resuspended in 1× PBS and stored at −20°C until further use.
Preparation of UBM
UBM was prepared from market-weight pigs (Tissue Source LLC, Lafayette, IN) as previously described (4). Briefly, the tunica serosa, muscularis externa, submucosa, and muscularis mucosa were removed by mechanical delamination, and the urothelial cells of the tunica mucosa were dissociated from the basement membrane by washing with deionized water. The remaining basement membrane and the lamina propria (collectively referred to as UBM) were decellularized by agitation in 0.1% peracetic acid with 4% ethanol for 2 hours at 300 rpm followed by PBS and type 1 water washes. UBM was then lyophilized and milled using a Wiley Mill with a #60 mesh screen.
Scanning electron microscopy
UBM was fixed in cold 2.5% glutaraldehyde for 24 hours followed by three 30-min washes in 1× PBS. Samples were then dehydrated in a graded series of alcohol (30, 50, 70, 90, and 100% ethanol) for 30 min per wash and then placed in 100% ethanol overnight at 4°C. Samples were washed three additional times in 100% ethanol for 30 min each and critical point–dried using a Leica EM CPD030 critical point dryer (Leica Microsystems, Buffalo Grove, IL, USA) with carbon dioxide as the transitional medium. Samples were then sputter-coated with a 4.5-nm-thick gold/palladium alloy coating using a 108 Auto sputter coater (Cressington Scientific Instruments, UK) and imaged with a JEOL JSM6330F scanning electron microscope (JEOL, Peabody, MA, USA).
Transmission electron microscopy
TEM imaging was conducted on MBV or liquid-phase EV loaded on carbon-coated grids and fixed in 4% paraformaldehyde as previously described (4). Grids were imaged at 80 kV with a JEOL 1210 TEM with a high-resolution Advanced Microscopy Techniques digital camera. The size of MBV was determined from representative images using JEOL TEM software.
Nanoparticle tracking analysis
Particle size and concentration of the liquid-phase EV and the MBV were calculated using a NanoSight (NS300) instrument equipped with fast video capture and particle-tracking software. Samples were diluted 1:500 to a final volume of 1000 μl using particle-free water. A syringe pump was used to dispense the sample into the system. Measurements were performed from three captures of 45 s each sample. For the video processing and particle calculation, the detection threshold was adjusted to 4. Data are presented as concentration versus particle size for each of the evaluated samples.
RNA isolation
Total RNA was isolated from 3T3 cells, liquid-phase EV, and MBV using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Before RNA isolation, liquid-phase EV and MBV samples were treated with ribonuclease A (10 μg/ml) at 37°C for 30 min to degrade any contaminating RNA. RNA quantity was determined using a NanoDrop spectrophotometer, and its quality was determined by Agilent Bioanalyzer 2100 (Agilent Technologies).
RNA-seq and bioinformatic analysis
The miRNA library preparation was initiated with 100 ng of each sample and the QIAseq miRNA Library Kit (Qiagen) following the manufacturer’s instructions. Briefly, mature miRNAs were ligated to adapters on their 3′ and 5′ ends. The ligated miRNAs were then reverse-transcribed to complementary DNA (cDNA) using a RT primer with unique molecular indices. The cDNA was then cleaned up to remove adapter primers, followed by amplification of the library with a universal forward primer and one of 48 reverse primers that assign a sample index. A presequencing quality control was performed using the Agilent RNA ScreenTape System. Next-generation sequencing was performed on a NextSeq 500 instrument with a loading concentration of 2.5 pM. Bioinformatic analysis was conducted by Genevia Technologies (Tampere, Finland). The quality of the sequencing reads was inspected using FastQC software. TrimGalore! (version 0.4.5) was used to remove the adapter sequences, with default settings, on all the samples. All reads were shortened to 21 bases, the typical size of miRNAs, using the fastx_trimmer software (FASTX-Toolkit by Hannon Lab, version 0.0.14). The reads of each sample were then aligned against the corresponding reference genome (hg38, GRCm38). Tables of miRNA counts across samples were created using the softwares bowtie (version 1.2.2) and miRDeep2 (version 0.0.8). In this process, precursor miRNA and mature miRNA sequences for each species involved in the study were taken from miRbase. Counts of mature miRNAs were obtained by taking the median of all precursor miRNAs associated with them. The counts of mature miRNAs of all samples were normalized using DESeq2. To ensure data quality before further analyses, PCA was performed and the results were visualized using ggplot2, separately for murine and human samples. Normalization of mature miRNA data and statistical testing between sample groups were performed with DESeq2. P values were corrected for multiple testing using the Benjamini-Hochberg method. miRNAs with adjusted P value of <0.05 and absolute log2 fold change >1 were considered as significantly differentially expressed. Tables of differentially expressed miRNAs were annotated with their targets and their confidences using the mirTARbase database of experimentally tested miRNA target interactions. Differentially expressed miRNAs were also annotated with predicted targets using the R package miRNAtap. miRNAtap aggregates the miRNA target predictions from five different databases (PicTar, DIANA, TargetScan, miRanda, and miRDB) and calculates an overall miRNA target score. The minimum amount of database sources required for a potential miRNAtarget interaction to be included into the annotations was 3.
Ingenuity pathway analysis
IPA software (version 01-14) was used for functional analysis of differentially expressed miRNAs. miRNA targets were identified using the IPA Core Analysis. The filter was set to Experimentally Observed findings to obtain information about significantly enriched molecular and cellular functions and physiological system development functions that were affected by the miRNAs.
qPCR validation
RT and qPCR were performed using the TaqMan Advanced miRNA Assays Protocol (Applied Biosystems). Briefly, 10 ng of total RNA was used with the TaqMan Advanced miRNA cDNA Synthesis Kit (Applied Biosystems, catalog no. A28007) to synthesize and adapt a 3′-poly(A) tail to the miRNAs. Universal RT primers recognizing the poly(A) tail were used to synthesize the cDNA in the RT reaction, followed by a miR-AMP step, using miR-AMP forward and reverse universal primers, to increase the number of cDNA molecules. The qPCR was made on a QuantStudio system machine using the TaqMan Fast Advanced Master Mix (Applied Biosystems, catalog no. 4444556) and specific TaqMan Advanced miRNA Assays (Applied Biosystems, catalog no. A25576) recognizing mmu-miR-163-5p, mmu-miR-27a-5p, mmu-miR-92a-1-5p, mmu-miR-451a, mmu-miR-93-5p, and mmu-miR-99b-5p. Fold change expression on the MBV sample was calculated for each of the specific targets using liquid-phase EV as a reference.
Immunoblot and silverstain assays
Liquid-phase EV and MBV, derived from three separate cultures of 3T3 fibroblasts, were respectively pooled and quantified by nanotracking particle analysis. For both immunoblot and silverstain analysis, an equal number of vesicles for both the liquid-phase EV and MBV samples were loaded onto the gel. MBV or liquid-phase EV (21 × 1011) was mixed with 2× Laemmli buffer (R&D Systems) containing 5% β mercaptoethanol (Sigma-Aldrich), resolved on a 4 to 20% gradient SDS–polyacrylamide gel electrophoresis (Bio-Rad), and then transferred onto a polyvinylidene difluoride membrane. Membranes were incubated overnight with the following primary antibodies: rabbit anti-CD63, rabbit anti-CD81, rabbit anti-CD9, and rabbit anti-Hsp70, at 1:1000 dilution (System Biosciences). Membranes were washed three times for 15 min each before and after they were incubated with goat anti-rabbit secondary antibody, at 1:5000 dilution (System Biosciences). The washed membranes were exposed to chemiluminescent substrate (Bio-Rad) and then visualized using a ChemiDoc Touch instrument (Bio-Rad). Silver staining of gels was performed using the Silver Stain Plus Kit (Bio-Rad) according to the manufacturer’s instruction and visualized using a ChemiDoc Touch instrument (Bio-Rad).
LC-MS analysis of phospholipids
Lipids were extracted from 3T3 cells, exosomes, and MBV by Folch procedure (45). MS analysis of phospholipids and their oxygenated products was performed on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific), as previously described (46). Briefly, phospholipids were separated on a normal-phase column [Luna 3 μm Silica (2) 100 Å, 150 × 2.0 mm (Phenomenex)] at a flow rate of 0.2 ml/min on a Dionex Ultimate 3000 HPLC system. The column was maintained at 35°C. The analysis was performed using gradient solvents (A and B) containing 10 mM ammonium acetate. Solvent A contained propanol:hexane:water (285:215:5, v/v/v), and solvent B contained propanol:hexane:water (285:215:40, v/v/v). All solvents were LC-MS grade. The column was eluted for 0 to 23 min with a linear gradient from 10 to 32% B, 23 to 32 min using a linear gradient of 32 to 65% B, 32 to 35 min with a linear gradient of 65 to 100% B, 35 to 62 min held at 100% B, and 62 to 64 min with a linear gradient from 100 to 10% B followed by an equilibration from 64 to 80 min at 10% B. Spectra were acquired in negative ion mode. Deuterated phospholipids were used as internal standards (Avanti Polar Lipids). Three technical replicates for each sample were run to evaluate reproducibility. Analysis of LC-MS data was performed using the software package Compound Discoverer (Thermo Fisher Scientific) with an in-house generated analysis workflow and nonoxidized/oxidized phospholipid database. Lipids were further filtered by retention time and confirmed by fragmentation mass spectrum.
LC-MS analysis of free fatty acids and their oxidation products
Free fatty acids were analyzed by LC-MS using a Dionex Ultimate 3000 HPLC system coupled online to a Q Exactive hybrid quadrupole-orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA), as previously described (47). Briefly, fatty acids and their oxidative derivatives were separated by a C18 column (Acclaim PepMap RSLC, 300 μm 15 cm, Thermo Fisher Scientific) using gradient solvents A [methanol (20%)/water (80%) (v/v)] and B [methanol (90%)/water (10%) (v/v)], both containing 5 mM ammonium acetate. The column was eluted at a flow rate of 12 μl/min using a linear gradient from 30% solvent B to 95% solvent B over 70 min, held at 95% B from 70 to 80 min, followed by a return to initial conditions by 83 min and re-equilibration for an additional 7 min. Spectra were acquired in negative ion mode. Analytical data were acquired and analyzed using Xcalibur software. A minimum of three technical replicates for each sample was run to increase the reproducibility.
Supplementary Material
Acknowledgments
We gratefully acknowledge T. Block and S. Griffey from StemBioSys for providing the ECM plates and K.-L. Aho from Genevia for the bioinformatic analysis. This project used the University of Pittsburgh HSCRF Genomics Research Core. We thank H. Deborah for conducting the small RNA-seq assay. Funding: This work was supported by NIH (HL114453-06, U19AI068021) and by Russian academic excellence program “5-100.” S.F.B. and G.S.H. were supported, in part, by NIH R01AR073527, “Mechanisms of functional skeletal muscle repair: Critical role of matrix associated IL-33.” Author contributions: G.S.H., V.E.K., and S.F.B. conceptualized and designed the research. G.S.H., C.P.M., M.C.C., Y.Y.T., V.A.T., Y.C.L., S.O.E.-M., M.H.M., and P.S.T. performed the experiments. G.S.H., C.P.M., Y.Y.T., V.A.T., Y.C.L., P.S.T., V.E.K., and S.F.B. analyzed the data and interpreted the results of experiments. G.S.H., C.P.M., Y.Y.T., and V.E.K. prepared the figures. G.S.H., V.E.K., and S.F.B. drafted the manuscript. Competing interests: S.F.B. is the chief scientific officer and equity holder in ECM Therapeutics Inc., which has license rights to MBV technology from the University of Pittsburgh. S.F.B. and G.S.H. are inventors on several patents related to this work filed by the University of Pittsburgh. The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data are available from the authors upon request.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/12/eaay4361/DC1
Table S1. Contents of CL, PA, PG and BMP in MBV, exosomes, and parent 3T3 cells.
REFERENCES AND NOTES
- 1.Baylón K., Rodríguez-Camarillo P., Elías-Zúñiga A., Díaz-Elizondo J. A., Gilkerson R., Lozano K., Past, present, and future of surgical meshes: A review. Membranes 7, E47 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hussey G. S., Dziki J. L., Badylak S. F., Extracellular matrix-based materials for regenerative medicine. Nat. Rev. Mater. 3, 159–173 (2018). [Google Scholar]
- 3.van Niel G., D’Angelo G., Raposo G., Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Huleihel L., Hussey G. S., Naranjo J. D., Zhang L., Dziki J. L., Turner N. J., Stolz D. B., Badylak S. F., Matrix-bound nanovesicles within ECM bioscaffolds. Sci. Adv. 2, e1600502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzpatrick L. E., McDevitt T. C., Cell-derived matrices for tissue engineering and regenerative medicine applications. Biomater. Sci. 3, 12–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Merwe Y., Faust A. E., Sakalli E. T., Westrick C. C., Hussey G., Chan K. C., Conner I. P., Fu V. L. N., Badylak S. F., Steketee M. B., Matrix-bound nanovesicles prevent ischemia-induced retinal ganglion cell axon degeneration and death and preserve visual function. Sci. Rep. 9, 3482 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu B., Lee B. W., Nakanishi K., Villasante A., Williamson R., Metz J., Kim J., Kanai M., Bi L., Brown K., di Paolo G., Homma S., Sims P. A., Topkara V. K., Vunjak-Novakovic G., Cardiac recovery via extended cell-free delivery of extracellular vesicles secreted by cardiomyocytes derived from induced pluripotent stem cells. Nat. Biomed. Eng. 2, 293–303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao S.-C., Guo S.-C., Li M., Ke Q.-F., Guo Y.-P., Zhang C.-Q., Chitosan wound dressings incorporating exosomes derived from MicroRNA-126-overexpressing synovium mesenchymal stem cells provide sustained release of exosomes and heal full-thickness skin defects in a diabetic rat model. Stem Cells Transl. Med. 6, 736–747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García-Manrique P., Matos M., Gutiérrez G., Pazos C., Blanco-López M. C., Therapeutic biomaterials based on extracellular vesicles: Classification of bio-engineering and mimetic preparation routes. J. Extracell. Vesicles 7, 1422676 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lötvall J., Hill A. F., Hochberg F., Buzás E. I., Di Vizio D., Gardiner C., Gho Y. S., Kurochkin I. V., Mathivanan S., Quesenberry P., Sahoo S., Tahara H., Wauben M. H., Witwer K. W., Théry C., Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell Vesicles 3, 26913 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skotland T., Hessvik N. P., Sandvig K., Llorente A., Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 60, 9–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Z., Zhang X., Zhao C., Choi J., Shi J., Song K., Turk J., Ma Z. A., Protection of pancreatic β-cells by group VIA phospholipase A2-mediated repair of mitochondrial membrane peroxidation. Endocrinology 151, 3038–3048 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyurina Y. Y., St. Croix C. M., Watkins S. C., Watson A. M., Epperly M. W., Anthonymuthu T. S., Kisin E. R., Vlasova I. I., Krysko O., Krysko D. V., Kapralov A. A., Dar H. H., Tyurin V. A., Amoscato A. A., Popova E. N., Bolevich S. B., Timashev P. S., Kellum J. A., Wenzel S. E., Mallampalli R. K., Greenberger J. S., Bayir H., Shvedova A. A., Kagan V. E., Redox (phospho) lipidomics of signaling in inflammation and programmed cell death. J. Leukoc. Biol. , (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyurina Y. Y., Shrivastava I., Tyurin V. A., Mao G., Dar H. H., Watkins S., Epperly M., Bahar I., Shvedova A. A., Pitt B., Wenzel S. E., Mallampalli R. K., Sadovsky Y., Gabrilovich D., Greenberger J. S., Bayır H., Kagan V. E., “Only a life lived for others is worth living”: Redox signaling by oxygenated phospholipids in cell fate decisions. Antioxid. Redox Signal. 29, 1333–1358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mouchlis V. D., Dennis E. A., Phospholipase A2 catalysis and lipid mediator lipidomics. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864, 766–771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rouzer C. A., Marnett L. J., Mechanism of free radical oxygenation of polyunsaturated fatty acids by cyclooxygenases. Chem. Rev. 103, 2239–2304 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Kuhn H., Banthiya S., van Leyen K., Mammalian lipoxygenases and their biological relevance. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1851, 308–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlame M., Greenberg M. L., Biosynthesis, remodeling and turnover of mitochondrial cardiolipin. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 3–7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braverman N. E., Moser A. B., Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta 1822, 1442–1452 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Glaser P. E., Gross R. W., Plasmenylethanolamine facilitates rapid membrane fusion: A stopped-flow kinetic investigation correlating the propensity of a major plasma membrane constituent to adopt an HII phase with its ability to promote membrane fusion. Biochemistry 33, 5805–5812 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Han X., Gross R. W., Plasmenylcholine and phosphatidylcholine membrane bilayers possess distinct conformational motifs. Biochemistry 29, 4992–4996 (1990). [DOI] [PubMed] [Google Scholar]
- 22.Rog T., Koivuniemi A., The biophysical properties of ethanolamine plasmalogens revealed by atomistic molecular dynamics simulations. Biochim. Biophys. Acta 1858, 97–103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ray R., Rai V., Lysophosphatidic acid converts monocytes into macrophages in both mice and humans. Blood 129, 1177–1183 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Tager A. M., LaCamera P., Shea B. S., Campanella G. S., Selman M., Zhao Z., Polosukhin V., Wain J., Karimi-Shah B. A., Kim N. D., Hart W. K., Pardo A., Blackwell T. S., Xu Y., Chun J., Luster A. D., The lysophosphatidic acid receptor LPA1 links pulmonary fibrosis to lung injury by mediating fibroblast recruitment and vascular leak. Nat. Med. 14, 45–54 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Masuda K., Haruta S., Orino K., Kawaminami M., Kurusu S., Autotaxin as a novel, tissue-remodeling-related factor in regressing corpora lutea of cycling rats. FEBS J. 280, 6600–6612 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Hines K. M., Ashfaq S., Davidson J. M., Opalenik S. R., Wikswo J. P., McLean J. A., Biomolecular signatures of diabetic wound healing by structural mass spectrometry. Anal. Chem. 85, 3651–3659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boilard E., Extracellular vesicles and their content in bioactive lipid mediators: More than a sack of microRNA. J. Lipid Res. 59, 2037–2046 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serhan C. N., Novel lipid mediators and resolution mechanisms in acute inflammation: To resolve or not? Am. J. Pathol. 177, 1576–1591 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lässer C., Jang S. C., Lötvall J., Subpopulations of extracellular vesicles and their therapeutic potential. Mol. Aspects Med. 60, 1–14 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Rilla K., Mustonen A. M., Arasu U. T., Härkönen K., Matilainen J., Nieminen P., Extracellular vesicles are integral and functional components of the extracellular matrix. Matrix Biol. 75-76, 201–219 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Xu L., Liu Y., Sun Y., Wang B., Xiong Y., Lin W., Wei Q., Wang H., He W., Wang B., Li G., Tissue source determines the differentiation potentials of mesenchymal stem cells: A comparative study of human mesenchymal stem cells from bone marrow and adipose tissue. Stem Cell Res. Ther. 8, 275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolhe R., Hunter M., Liu S., Jadeja R. N., Pundkar C., Mondal A. K., Mendhe B., Drewry M., Rojiani M. V., Liu Y., Isales C. M., Guldberg R. E., Hamrick M. W., Fulzele S., Gender-specific differential expression of exosomal miRNA in synovial fluid of patients with osteoarthritis. Sci. Rep. 7, 2029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eitan E., Green J., Bodogai M., Mode N. A., Bæk R., Jørgensen M. M., Freeman D. W., Witwer K. W., Zonderman A. B., Biragyn A., Mattson M. P., Noren Hooten N., Evans M. K., Age-related changes in plasma extracellular vesicle characteristics and internalization by leukocytes. Sci. Rep. 7, 1342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao Z.-y., Chen W.-b., Shao S.-s., Ma S.-z., Yang C.-b., Li M.-z., Zhao J.-j., Gao L., Role of exosome-associated microRNA in diagnostic and therapeutic applications to metabolic disorders. J. Zhejiang Univ. Sci. B 19, 183–198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gimona M., Pachler K., Laner-Plamberger S., Schallmoser K., Rohde E., Manufacturing of human extracellular vesicle-based therapeutics for clinical use. Int. J. Mol. Sci. 18, E1190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willis G. R., Kourembanas S., Mitsialis S. A., Toward exosome-based therapeutics: Isolation, heterogeneity, and fit-for-purpose potency. Front. Cardiovasc. Med. 4, 63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai R. C., Tan S. S., Yeo R. W. Y., Choo A. B. H., Reiner A. T., Su Y., Shen Y., Fu Z., Alexander L., Sze S. K., Lim S. K., MSC secretes at least 3 EV types each with a unique permutation of membrane lipid, protein and RNA. J. Extracell. Vesicles 5, 29828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.György B., Szabó T. G., Pásztói M., Pál Z., Misják P., Aradi B., László V., Pállinger É., Pap E., Kittel Á., Nagy G., Falus A., Buzás E. I., Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell. Mol. Life Sci. 68, 2667–2688 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuster-Matanzo A., Gessler F., Leonardi T., Iraci N., Pluchino S., Acellular approaches for regenerative medicine: On the verge of clinical trials with extracellular membrane vesicles? Stem Cell Res. Ther. 6, 227 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.György B., Hung M. E., Breakefield X. O., Leonard J. N., Therapeutic applications of extracellular vesicles: Clinical promise and open questions. Annu. Rev. Pharmacol. Toxicol. 55, 439–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y., Puperi D. S., Grande-Allen K. J., West J. L., Ascorbic acid promotes extracellular matrix deposition while preserving valve interstitial cell quiescence within 3D hydrogel scaffolds. J. Tissue Eng. Regen. Med. 11, 1963–1973 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X.-D., Dusevich V., Feng J. Q., Manolagas S. C., Jilka R. L., Extracellular matrix made by bone marrow cells facilitates expansion of marrow-derived mesenchymal progenitor cells and prevents their differentiation into osteoblasts. J. Bone Miner. Res. 22, 1943–1956 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Lai Y., Sun Y., Skinner C. M., Son E. L., Lu Z., Tuan R. S., Jilka R. L., Ling J., Chen X.-D., Reconstitution of marrow-derived extracellular matrix ex vivo: A robust culture system for expanding large-scale highly functional human mesenchymal stem cells. Stem Cells Dev. 19, 1095–1107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shelke G. V., Lässer C., Gho Y. S., Lötvall J., Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J. Extracell. Vesicles 3, 24783 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Folch J., Lees M., Sloane Stanley G., A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 (1957). [PubMed] [Google Scholar]
- 46.Tyurina Y. Y., Kisin E. R., Murray A., Tyurin V. A., Kapralova V. I., Sparvero L. J., Amoscato A. A., Samhan-Arias A. K., Swedin L., Lahesmaa R., Fadeel B., Shvedova A. A., Kagan V. E., Global phospholipidomics analysis reveals selective pulmonary peroxidation profiles upon inhalation of single-walled carbon nanotubes. ACS Nano 5, 7342–7353 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tyurina Y. Y., Poloyac S. M., Tyurin V. A., Kapralov A. A., Jiang J., Anthonymuthu T. S., Kapralova V. I., Vikulina A. S., Jung M. Y., Epperly M. W., Mohammadyani D., Klein-Seetharaman J., Jackson T. C., Kochanek P. M., Pitt B. R., Greenberger J. S., Vladimirov Y. A., Bayır H., Kagan V. E., A mitochondrial pathway for biosynthesis of lipid mediators. Nat. Chem. 6, 542–552 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/12/eaay4361/DC1
Table S1. Contents of CL, PA, PG and BMP in MBV, exosomes, and parent 3T3 cells.


