Fig. 5. Top DG compounds affect cellular redox balance.
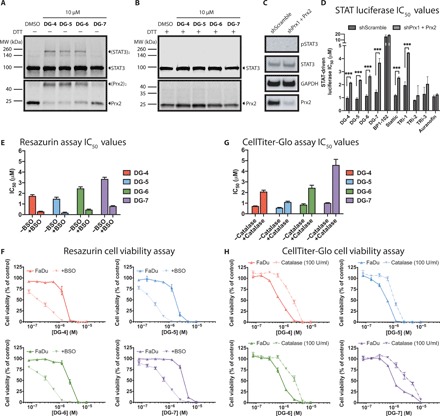
(A) Western blot analyses of HEK293 cells treated with the top DG compounds. Following a 30-min exposure to compounds, STAT3 and Prx2 were oxidized to form dimers in the absence of reducing agents such as DTT. (B) Similar to the experiment shown in (A), however, the addition of a reducing agent (DTT) during protein sample preparation reduces the inter- and intraprotein disulfide interactions, eliminating the bands corresponding to oligomeric STAT3 and Prx2. (C) Western blot analyses of pSTAT3/STAT3 and Prx2 expression in HEK293-shScramble and HEK293–shPrx1 + Prx2 cells. (D) IC50 values of indicated compounds for STAT-driven luciferase inhibition curves in HEK293-shScramble and HEK293–shPrx1 + Prx2 cells stimulated with IL6 (50 ng/ml) and sIL6R (100 ng/ml). Prx1 + Prx2 knockdown could rescue STAT-dependent transcription leading for our top DG compounds and for TRi-1 and Stattic. (E) Resazurin viability IC50 values for FaDu cells incubated with top DG compounds for 72 hours. Cells were preincubated with or without buthionine sulfoximine (BSO) (100 μM) for 24 hours before the addition of top DG compounds. (F) Cell viability curves for the data described in (E). (G) CellTiter-Glo viability IC50 values for FaDu cells incubated with top DG compounds for 72 hours. Cells were preincubated with or without catalase (100 U/ml) for 4 hours before the addition of top inhibitors. (H) Cell viability curves for the data described in (G).
