Abstract
Aim
The X-ray repair cross-complementing (XRCC) gene polymorphisms influence esophageal carcinogenesis by altering the DNA repair capacity. The present study was designed to screen five single nucleotide polymorphisms (SNPs) of XRCC genes for their susceptibility to esophageal cancer (EC) risk. There is no previous report on these polymorphisms for EC from India, where EC frequency is high.
Methods
The present study included 497 subjects (213 EC patients and 284 healthy controls). The polymorphisms were screened using the PCR-RFLP method and allele and genotype distribution were compared using chi-square test. Association analysis was done by haplotype analysis and linkage disequilibrium (LD) analysis. Gene–gene interactions were identified using multifactor dimensionality reduction (MDR). The risk was calculated using binary logistic regression.
Results
For XRCC1 p.Arg399Gln, a decreased risk for EC was associated with the AA genotype [OR (95% CI): 0.53 (0.3–0.95), p=0.03] even after adjusting for various covariates [OR (95% CI): 0.49 (0.26–0.9), p=0.024] and with the recessive model [OR (95% CI): 0.49 (0.27–0.8), p=0.016]. The GA genotype of p.Arg280His was associated with an increased risk for EC [OR (95% CI): 1.7 (1.0–2.82), p= 0.045] after adjustments. The two XRCC1 polymorphisms, p.Arg399Gln and p.Arg194Trp were in slight LD among EC patients (D̍́=0.845, r2=0.042). XRCC2 and XRCC3 polymorphisms were not associated with EC risk.
Conclusion
XRCC1 p.Arg399Gln plays a protective role in the development of the EC. The study is the first report from India, providing baseline data about genetic polymorphisms in DNA repair genes XRCC1, XRCC2 and XRCC3 modulating overall EC risk.
Keywords: XRCC1, XRCC2, XRCC3, polymorphisms, esophageal cancer
Introduction
Esophageal cancer (EC) ranks as the sixth leading cause of death from cancer constituting 7% of all gastrointestinal cancers.1 In India, EC is the fourth most common cause of deaths related to cancer and the second most common cancer among men and fourth among women.2 Esophageal carcinogenesis is a complex multistep process with poor prognosis where environmental, geographic and genetic factors appear to play a major role.3 Cancer is a disease characterized by the failure of DNA repair mechanisms. XRCC1 protein as a part of Base excision repair (BER) pathway plays an efficient role in repairing DNA single-strand breaks.4 It is encoded by the XRCC1 gene located on 19q13.2, comprising of 17 exons and 16 introns.5 Two XRCC1 polymorphisms, p.Arg194Trp (exon 6) and p. Arg280His (exon 9) affect the function of the protein.6 XRCC1 p.Arg399Gln polymorphism in exon 107 has been associated with breast,8 lung9 and head and neck cancers.10
XRCC2 (chromosome location 7q36.1) plays a major role in Homologous recombination repair (HRR) pathway.11 XRCC2 p.Arg188His polymorphism located in exon312 has been associated with cancers like pancreatic,13 ovarian,14 oral15 and upper aerodigestive tract cancers.16
XRCC3 (chromosome location 14q32.3) encodes a 346 amino acid polypeptide17 that participates in DNA double-strand break repair. Variation in expression of XRCC3 has been reported in various cancers, like gastric, breast, lung, skin and colorectal.18–21 The most common polymorphism in XRCC3 p.Thr241Met in exon 7 can influence the ability to repair DNA.21
Allelic variants of XRCC1, XRCC2, and XRCC3 are associated with risk of different types of cancer among different populations all over the world.22–24 In the world’s second-most populous country India, the large and diverse ethnic subpopulations25 need to be studied to understand the genetic basis of several region-specific complex disorders. To the best of our knowledge, studies related to these DNA-repair genes are very few for esophageal cancer in India (Table 1). The state of Punjab, India, has very high age-adjusted rate (AAR) for EC; it ranks second in males after prostate cancer and fourth after breast, ovarian and cervical cancer in females in this region.26,27
Table 1.
Studies Showing Association of XRCC1, XRCC2 and XRCC3 (p.Thr241Met) Polymorphism in Different Cancers in Populations from Different Geographical Regions of India
| Authors | Population of Region (State) | SNPs | Disease | Number of Subjects | Results |
|---|---|---|---|---|---|
| GIT Cancers | |||||
| Present Study | North India (Punjab) |
XRCC1 p.Arg399Gln, p.Arg194Trp and p.Arg280His XRCC2 p.Arg188His XRCC3 p.Thr241Met |
Esophageal Cancer |
497 | AA genotype of XRCC1 p.Arg399Gln associated with reduced ESCC risk |
| 34* | North India (Punjab/Chandigarh) |
XRCC1 p.Arg399Gln | Esophageal Squamous Cell Carcinoma | 280 | AA genotype associated with reduced ESCC risk |
| 43* | North India(Uttar pradesh) |
XRCC1 p.Arg399Gln p.Arg194His |
Gallbladder Cancer | 377 | GA and AA genotype of p.Arg399Gln associated with decreased risk. No association of p.Arg194Trp polymorphism |
| 50 | North India (Kashmir) | XRCC1 p.Arg399Gln | Colorectal Cancer | 260 | Association of GA genotype with increased cancer risk |
| 44* | North India (Kashmir) | XRCC1 p.Arg399Gln | Colorectal Cancer | 266 | Association of AA genotype with decreased risk |
| 52 | Eastern India (West Bengal) |
XRCC1- p.Arg399Gln, p.Arg194Trp and p.Arg280His | Gastric Cancer | 152 | GA and GG genotype of p.Arg399Gln associated with increased cancer risk No association of p.Arg194Trp and p.Arg280His polymorphism |
| 72* | South India | XRCC1 p.Arg194Trp | Gastric Cancer | 600 | No association |
| Cancers of Other Sites | |||||
| Eastern India |
XRCC1 p.Arg399Gln and p.Arg194Trp XRCC3 p.Thr241Met |
Oral submucous fibrosis (OSF) | 188 | No association of XRCC1 p.Arg399Gln and p.Arg194trp polymorphism CT genotype of XRCC3 p.Thr241Met associated with increased risk of OSF |
|
| 46* | North India | XRCC1 p.Arg194Trp, p.Arg280His and p.Arg399Gln | Hepatocellular Carcinoma | 407 | TT of p.Arg280His associated with increased risk. AA genotype of p.Arg399Gln associated with decreased risk |
| 90 | North eastern India |
XRCC1 p.Arg194Trp XRCC3 p.Thr241Met |
Breast Cancer | 998 | Arg/Trp and Trp/Trp genotype of XRCC1 p.Arg194Trp associated with an increased risk No association of XRCC3 p.Thr241Met |
| 101 | Western India (Maharashtra) |
XRCC1- p.Arg399Gln, p.Arg194His and p.Arg280His XRCC2- p.Arg188His XRCC3- p.Thr241Met |
Breast Cancer | 350 | No association of XRCC1 (p.Arg399Gln, p.Arg194His and p.Arg280His), XRCC2 (p.Arg188His) and XRCC3 (p.Thr241Met)with breast cancer |
| 55 | South India | XRCC1 p.Arg194Trp, p.Arg280His and p.Arg399Gln | Cervical Cancer | 250 | No association |
| 51 | North India | XRCC1 p.Arg399Gln | Prostate Cancer | 450 | Association of AA genotype with increased risk |
| 57 | South India | XRCC1 p.Arg194Trp, p.Arg280His and p.Arg399Gln | Lymphoblastic Leukemia | 234 | Association of Arg/Gln and Gln/Gln genotype of p.Arg399Gln with increased risk. No association for p.Arg280His and p.Arg194Trp polymorphism |
| 58 | South India | XRCC1 p.Arg399Gln | Acute Myeloid Leukemia | 422 | Association of GA- genotype with increased risk of AML |
| 102 | Middle India (Madhya Pradesh) | XRCC1 p.Arg194Trp, p.Arg280His and p.Arg399Gln | Sickle Cell Anemia | 500 | GA and AA genotype of p.Arg280His associated with an increased risk. No association of XRCC1 p.Arg194Trp and p.Arg399Gln polymorphisms |
| 45* | North India | XRCC1 p.Arg194Trp, p.Arg280His and p.Arg399Gln | Head and Neck Cancer | 556 | AA genotype of p.Arg399Gln and TT genotype of p. Arg194Trp associated with reduced risk. No association of p.Arg280Hispolymorphism. |
Note: *Studies with similar results as present study.
Therefore, the present study was carried out to explore the role of five polymorphisms of XRCC genes; XRCC1 (p.Arg399Gln, p.Arg194Trp, p.Arg280His), XRCC2 (p.Arg188His) and XRCC3 (p.Thr241Met) in the risk of EC in the population of Punjab, India.
Materials and Methods
Study Population and Sample Collection
The present case-control study was conducted after due ethical clearance according to the tenets of the Declaration of Helsinki by the institutional ethics committee of Guru Nanak Dev University, Amritsar. The study sample included 497 subjects: 213 (125 females and 88 males) esophageal cancer patients without a history of any other cancer, any treatment before sample collection and pre-operative from Sri Guru Ram Das Institute of Medical Sciences and Research, Amritsar. Complete information including age, gender, lifestyle, diet, family history, and environmental factors were recorded on a proforma after written informed consent from all the subjects. Two hundred eighty-four (187 females and 97 males), age and gender-matched unrelated healthy controls from the same geographical region were also included in the study (Table 2).
Table 2.
Clinical Characteristics of the Esophageal Cancer Patients and the Controls
| Characteristics | Cases (n=213) | Controls (n=284) | Odds Ratio (95% CI) | p-value |
|---|---|---|---|---|
| Age Below 55 55 or above Age(years) |
94 119 55.7±12.4 |
144 140 54.4±11 |
0.15 0.22 |
|
| Sex Females Males |
125 88 |
187 97 |
0.07 |
|
| Habitat Rural Urban |
178 35 |
241 43 |
1.14(0.67–1.70) |
0.70 |
| Diet Vegetarians Non-vegetarians |
108 105 |
142 142 |
0.97(0.68–1.40) |
0.87 |
| Smoking No Yes |
181 32 |
279 5 |
9.83(3.7–25.72) |
<0.0001 |
| Drinking No Yes |
153 60 |
233 51 |
1.79(1.17–2.14) |
0.007 |
| Family history No Yes |
192 21 |
284 0 |
||
| Type of cancer SCC ADC |
187 26 |
– – |
||
| Clinical Stage (I) (II) (III) IV |
10 67 122 14 |
– – – – |
Notes: p-value less than 0.05 taken as statistically significant (bold values).
Abbreviation: SCC, squamous cell carcinoma, ADC, adenocarcinoma.
Collection of Blood samples and genotyping: Intravenous blood sample was collected in EDTA from each subject. Genomic DNA from blood was collected using standard Phenol chloroform method.28 Previously published primer sequences for XRCC1 p.Arg399Gln,29 XRCC1 p.Arg194Trp,24 XRCC1 p.Arg280His,22 XRCC2 p.Arg188His30 and XRCC3 p.Thr241Met23 polymorphisms were used to amplify the target region. The total volume of the PCR reaction mixture was 15µL containing the reagents: 50ng of template DNA, 10pmol of each primer (Sigma, St. Louis, MO, USA), 10X PCR buffer with 1.5 mM MgCl2, 50 μM of dNTPs mix and 0.6 units of Taq polymerase (Bangalore GeNei).
The PCR products were digested with appropriate restriction endonucleases using manufacturer recommendations (NEB, UK) and analyzed on ethidium bromide stained agarose gel to obtain genotypes based on the size of fragment obtained (Table 3). To reassure of the quality of the result 10% of the samples were re-genotyped.
Table 3.
Details of Amplification and Digestion Conditions Used for Genotyping of XRCC1, XRCC2 and XRCC3 Polymorphisms
| Polymorphism (Ref SNP) |
Location | Nucleotide Change |
Annealing Temperature |
Amplicon Size (bp) |
Restriction Enzyme |
Size of Digested Fragments |
|---|---|---|---|---|---|---|
|
XRCC1 p.Arg194Trp (rs1799782) |
Exon 6 | C>T | 59°C | 491 | MspI | CC(292,174) CT(313,292,174) TT(313,174) |
| p.Arg280His (rs25489) |
Exon 9 | G>A | 66°C | 861 | RsaI | GG(597,201,63) GA(660,597,201,63) AA(660,201) |
| p.Arg399Gln (rs25487) |
Exon 10 | G>A | 59°C | 615 | MspI | GG(374,241) GA(615,374,241) AA(615) |
|
XRCC2 p.Arg188His (rs3218536) |
Exon 3 | G>A | 59°C | 290 | HphI | GG(290) GA(290,148,142) AA(148,142) |
|
XRCC3 p.Thr241Met (rs1625895) |
Exon 7 | C>T | 62°C | 552 | NlaIII | CC(313,239) CT(313,239,208,10) TT (239,208,105) |
Abbreviation: SNP, single nucleotide polymorphism.
Statistical Analysis
Statistical analysis was done using SPSS software version 20 (SPSS, Chicago, IL, USA). The analysis of categorical variables was done using the chi-square test and the continuous independent variables were expressed as mean ± standard deviation using the Student’s t-test. Hardy-Weinberg equilibrium was tested using the chi-square test. Binary logistic regression analysis was done to measure the risk of EC with genotypes of selected polymorphisms and the values were adjusted for the possible confounding effect of age, gender, diet, smoking and alcohol consumption. Odds ratios (ORs), p-values and 95% confidence interval (CI) were calculated as the measure of the strength of association of the SNPs and esophageal cancer. Haplotypes and linkage disequilibrium patterns were studied using Haploview software. Furthermore, multifactor dimensionality reduction (MDR) factor analysis version 2 was used to calculate the effect of Gene–gene interactions on esophageal cancer.
Results
The detailed characteristics of 497 subjects (213 EC patients and 284 controls) are given in Table 2 and the allele and genotype frequencies are summarised in Table 4 and Table 5. All the studied subjects were residents of Punjab, India. The endogamous caste groups were Jatt Sikh, Khatri and Scheduled Caste. The subjects were ethnic Punjabi with Scythian and Caucasian racial admixture.31 In the present study 5 polymorphisms; XRCC1 (p.Arg399Gln, p.Arg280His, p.Arg194Trp), XRCC2 (p.Arg188His) and XRCC3 (p.Thr241Met) were studied for the association with risk of esophageal cancer (EC). For XRCC1 p.Arg280His polymorphism, A allele was associated with an increased risk of EC (OR=1.60, 95% CI= 1.02–2.52, p=0.04). According to the Bivariate logistic regression for stratification, genotypes GA and GG+GA (OR=1.8,95% CI =1.1–2.9, p=0.025) provide ~2 fold increased risk of EC after adjusting the confounding effect of age, gender, diet pattern, smoking and alcohol consumption. For XRCC1p.Arg399Gln polymorphism, AA genotype (OR= 0.53, 95% CI=0.3–0.95, p=0.03) was associated with ~50% decreased the risk of EC. This association persisted and became stronger (OR=0.5, 95% CI=0.3–0.9, p=0.024) after adjustments to age, sex, diet pattern, smoking and alcohol consumption. Smoking and Alcohol intake was reported only by male subjects. Smoking was not reported by many subjects probably because of prevalent socio-religious beliefs in the region prohibit smoking and tobacco intake and alcohol intake. No significant difference in allele (p=0.54) and genotype (p=0.52) frequencies of XRCC1 p.Arg194Trp polymorphism was observed. No association with EC risk was observed for XRCC2 p.Arg188His and XRCC3 p.Thr241Met polymorphism (p=0.53) in the subjects.
Table 4.
Allele and Genotype Frequencies of XRCC1, XRCC2 and XRCC3 Gene Polymorphisms and Their Association with Esophageal Cancer Risk
| Gene | SNP | Allele | Allele Frequency | Odds Ratio (95% CI) |
p value | Genotype | Cases n (%) | Controls n(%) | Chi Square (ᵡ2) | p value* | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases (n%) |
Controls (n%) |
||||||||||
| XRCC1 | rs25489 p.Arg280His |
G A |
382(89.7) 44(10.3) |
530(93.3) 38(6.7) |
Reference 1.60(1.02–2.51) |
0.04 | GG GA AA |
171(80.3) 40(18.8) 2(0.9) |
246(86.6) 38(13.4) 0(0) |
4.9 | 0.08 |
| rs25487 p.Arg399Gln |
G A |
283(66.5) 143(33.5) |
344(60.6) 224(39.4) |
Reference 0.78 (0.6–1.0) |
0.06 | GG GA AA |
92(43.2) 99(46.5) 22(10.3) |
109(38.4) 126(44.4) 49(17.2) |
5.51 | 0.06 | |
| rs1799782 p.Arg194Trp |
C T |
394(92.5) 32(7.5) |
531(93.5) 37(6.5) |
Reference 1.16(0.71–1.92) |
0.54 | CC CT TT |
181(85) 32(15) 0(0) |
247(87) 37(13) 0 |
0.41 | 0.81 | |
| XRCC2 | rs3218536 p.Arg188His |
G A |
388(91.1) 38(8.9) |
515(90.7) 52((9.3) |
Reference 0.97(0.62–1.51) |
0.89 | GG GA AA |
175(82) 38(18) 0(0) |
232(81.7) 51(18) 1(0.3) |
0.75 | 0.68 |
| XRCC3 | rs1625895 p.Thr241Met |
C T |
334(78.4) 92(21.6) |
449(79) 119(21) |
Reference 1.03(0.76–1.43) |
0.80 | CC CT TT |
129(60.6) 76(35.7) 8(3.7) |
178(62.7) 93(32.7) 13(4.6) |
0.59 | 0.74 |
Notes: Wild type genotype was taken as reference. *Distribution of overall genotypic frequencies between cases and controls done by χ2 test (df, 2). P-value less than 0.05 taken as statistically significant (bold values).
Abbreviation: SNP, single nucleotide polymorphism.
Table 5.
Distributions of XRCC1, XRCC2 and XRCC3 Polymorphisms Genotypes and Development of Esophageal Cancer
| Gene | SNP | Genotype | Cases n(%) | Controls n(%) | Unadjusted OR(CI)(p) | AOR* (CI) (p value) | AOR** (CI) (p value) |
|---|---|---|---|---|---|---|---|
| XRCC1 | rs25489 p.Arg280His |
GG GA AA GA+AA |
171(80.3) 40(18.8) 2(0.9) |
246(86.6) 38(13.4) 0(0) |
Reference 1.5(0.9–2.4) p=0.09 NC 1.6(0.9–2.5) p=0.06 |
Reference 1.6(1.0–2.6) p=0.06 NC 1.6(1.0–2.6) p=0.04 |
Reference 1.7 (1.0–2.8) p=0.04 NC 1.8(1.1–2.9) p=0.02 |
| rs25487 p.Arg399Gln |
GG GA AA GA+AA |
92(43.2) 99(46.5) 22(10.3) |
109(38.4) 126(44.4) 49(17.2) |
Reference 0.9(0.6–1.3) p=0.7 0.5(0.3–0.9) p=0.03 0.8(0.6–1.2) p=0.28 |
Reference 0.9(0.6–1.4) p=0.7 0.5(0.3–0.9) p=0.03 0.8(0.6–1.2) p=0.27 |
Reference 1.0(0.7–1.5) p=0.9 0.5(0.3–0.9)p=0.024 0.9(0.6–1.3) p=0.4 |
|
| rs1799782 p.Arg194Trp |
CC CT TT |
181(85) 32(15) 0(0) |
247(87) 37(13) 0 |
Reference 1.2(0.7–1.9) p=0.52 NC |
Reference 1.2(0.7–2.1) p=0.4 NC |
Reference 1.1(0.7–2.0) p= 0.5 NC |
|
| XRCC2 | rs3218536 p.Arg188His |
GG GA AA GA+AA |
175(82) 38(18) 0(0) |
232(81.7) 51(18) 1(0.3) |
Reference 0.9(0.6–1.6) p=0.95 NC 0.9(0.6–1.5) p=0.89 |
Reference 1.0(0.6–1.6) p=0.9 NC 0.99(0.6–1.6) p=0.9 |
Reference 0.9(0.6–1.5) p=0.8 NC 0.9(0.5–1.5) p=0.7 |
| XRCC3 | rs1625895 p.Thr241Met |
CC CT TT CT+TT |
129(60.6) 76(35.7) 8(3.7) |
178(62.7) 93(32.7) 13(4.6) |
Reference 1.1(0.7–1.6) p=0.53 0.8(0.3–2.1) p=0.72 1.09(0.7–1.6) p=0.63 |
Reference 1.1(0.7–1.6) p=0.6 0.8(0.3–2.2) p=0.8 1.06(0.7–1.5) p=0.75 |
Reference 1.1(0.7–1.6) p=0.6 0.8(0.3–2.3) p=0.8 1.06(0.7–1.6) p=0.77 |
Notes: Wild type genotype is taken as reference, p<0.05 taken as statistically significant (bold values). AOR* Adjusted Odds Ratio for age, gender and diet pattern using Binary logistic regression. AOR** Adjusted Odds Ratio for age, gender, diet pattern, cigarette smoking and alcohol consumption using Binary logistic regression analysis.
Genetic model analysis of XRCC1 p.Arg399Gln and XRCC3p.Thr241Met (Table 6) revealed a decreased risk of EC under the recessive model AA vs GG+GA (OR=0.55, 95% CI=0.32–0.95, p=0.027) for XRCC1p.Arg399Gln polymorphism which became more significant after adjustment with binomial logistic regression (OR=0.49, 95% CI=0.27–0.88, p=0.016). For p.Thr241Met polymorphism no genotype combination was associated with EC.
Table 6.
Association Analysis of XRCC1 p.Arg399Gln and XRCC3p.Thr241Met Polymorphisms
| SNPs | Comparison | Unadjusted OR (CI) (p value) |
AOR* (CI) (p value) | AOR** (CI) (p value) |
|---|---|---|---|---|
|
XRCC1 p.Arg399Gln |
Dominant model (AA+GAvsGG) Codominant model(AAvsGA/GGvsGA) Recessive model (GG+GAvsAA) |
0.82(0.57–1.22) p=0.280 0.78 (0.60 −1.01) p=0.062 0.55(0.32–0.95) p=0.027 |
0.82(0.56–1.18) p=0.27 1.16(0.71–1.64) p=0.56 0.53(0.30–0.95) p=0.02 |
0.92(0.59–1.31) p=0.46 1.23(0.82–1.82) p=0.29 0.49(0.27–0.83) p=0.02 |
|
XRCC3 p.Thr241Met |
Dominant model (TT+CTvsCC) Codominant model(TTvsCT/TTvsCC) Recessive model (CC+CTvsTT) |
1.09(0.76–1.58) p=0.63 1.04 (0.76 −1.42) p=0.80 0.81(0.33–1.92) p=0.65 |
1.06(0.73–1.5) p=0.75 1.14(0.75–1.62) p=0.62 0.84(0.34–2.08) p=0.69 |
1.06(0.71–1.63) p=0.77 1.13(0.74–1.65) p=0.61 0.82(0.33–2.05) p=0.64 |
Notes: Wild type genotype is taken as reference, p<0.05 taken as statistically significant (bold values). AOR* Adjusted Odds Ratio for age, gender and diet pattern using binary logistic regression. AOR** Adjusted Odds Ratio for age, gender, diet pattern, cigarette smoking and alcohol consumption using binary logistic regression.
Haplotype and Linkage Disequilibrium Analysis
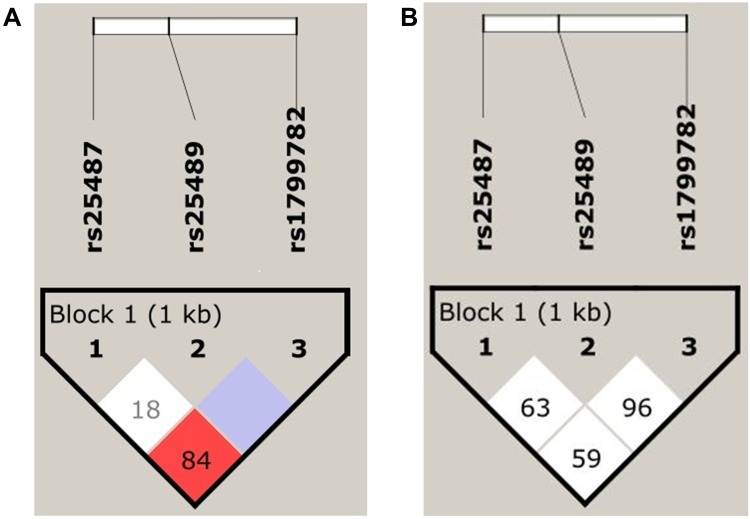
The haplotype frequencies of three polymorphisms of XRCC1 were examined for their association with EC risk (Table 7). Based on the measures of linkage disequilibrium (LD), the two polymorphisms of XRCC1gene, p.Arg399Gln and p.Arg194Trp, were in slight LD among EC patients (D̍́=0.845, r2=0.042) (Figure 1A). However, control subjects were not in LD for any of the polymorphism (Figure 1B). The haplotype GGT (p.Arg399Gln, p.Arg280His, and p.Arg194Trp) was predominant in EC cases as compared to controls, but the difference was statistically non-significant (p=0.1). The possible genotype combinations of three studied polymorphisms of XRCC1 were also examined (Table 8). The combinations comprising the AA genotype (p.Arg399Gln) occurred significantly more often in controls than patients, with AA-CC-CC combination associated with significantly decreased risk of EC (OR=0.5, 95% CI=0.29–0.91, p=0.020).
Table 7.
Haplotype Frequencies of XRCC1 Gene Polymorphisms and Their Association with the Risk of Esophageal Cancer
| Haplotype XRCC1 p.Arg399Gln-p.Arg280His-p.Arg194Trp | Studied Population % (n)(497) | Cases N | Controls | χ2 (p value)* | Odds Ratio (OR)(95% CI) p value** | |
|---|---|---|---|---|---|---|
| G-G-C | 0.501(249) | 0.508(108) | 0.496(141) | 0.1 (p=0.7) | Reference | |
| A-G-C | 0.347(174) | 0.314(67) | 0.372(107) | 3.6 (p=0.05) | 0.82(0.6–1.2) p=0.3 | |
| G-G-T | 0.075(37) | 0.096(20) | 0.058(17) | 5.0(p=0.02) | 1.5(0.7–3.1) p= 0.1 | |
| G-A-C | 0.055(27) | 0.060(13) | 0.052(14) | 0.3(p=0.5) | 1.2(0.5–2.7) p= 0.6 | |
| A-A-C | 0.014(7) | 0.015(3) | 0.014(4) | 0.02(p=0.8) | 0.9(0.2–4.4) p=0.9 | |
| A-G-T | 0.008(4) | 0.007(2) | 0.008(2) | 0.07(p=0.7) | 1.3(0.2–9.4) p=0.8 | |
| LD Measures | Esophageal Cancer Patients | |||||
| D̍̀ | r2 | D̀ | ||||
| p.Arg399Gln-p.Arg194Trp | 0.845 | 0.042 | 0.596 | |||
| p.Arg280His-p.Arg194Trp | 1.0 | 0.009 | 0.969 | |||
| p.Arg399Gln-p.Arg280His | 0.188 | 0.001 | 0.633 | |||
Notes: P* Comparison for esophageal cancer patients and controls for each haplotype. P** Comparison for esophageal cancer patients and controls in comparison to the wild type genotype taken as reference, P<0.05 taken as statistically significant (bold values).
Figure 1.
LD plot showing the position of the three XRCC1 polymorphisms and pair-wise D’ values observed in the study population with respect to (A) Esophageal Cancer (B) Controls.
Table 8.
Association of XRCC1 Genotype Combinations with Esophageal Cancer
| Genotype Combination* | Patients | Controls | Odds Ratio (95% CI) |
p value |
|---|---|---|---|---|
| AA-CC-CC | 19 | 45 | 0.5(0.3–0.9) | 0.02 |
| AA-nn-CC | 21 | 47 | 0.5(0.3–0.9) | 0.03 |
| AA-CC-nn | 20 | 47 | 0.5(0.3–0.9) | 0.02 |
| nn-CT-CC | 28 | 34 | 1.1(0.6–1.9) | 0.69 |
| An-Tn-CC | 18 | 20 | 1.2(0.6–2.4) | 0.56 |
| An-nn-Cn | 121 | 175 | 0.8(0.6–1.2) | 0.28 |
| nn-Tn-Cn | 32 | 37 | 1.2(0.7–1.9) | 0.52 |
| Gn-Tn-Cn | 30 | 35 | 1.2(0.7–1.9) | 0.56 |
Notes: *Genotypes of XRCC1 p.Arg399Gln, p.Arg194Trp and p.Arg280His. n: any allele. p value <0.05 is taken as statistically significant (bold values)
Interaction Analysis (MDR)
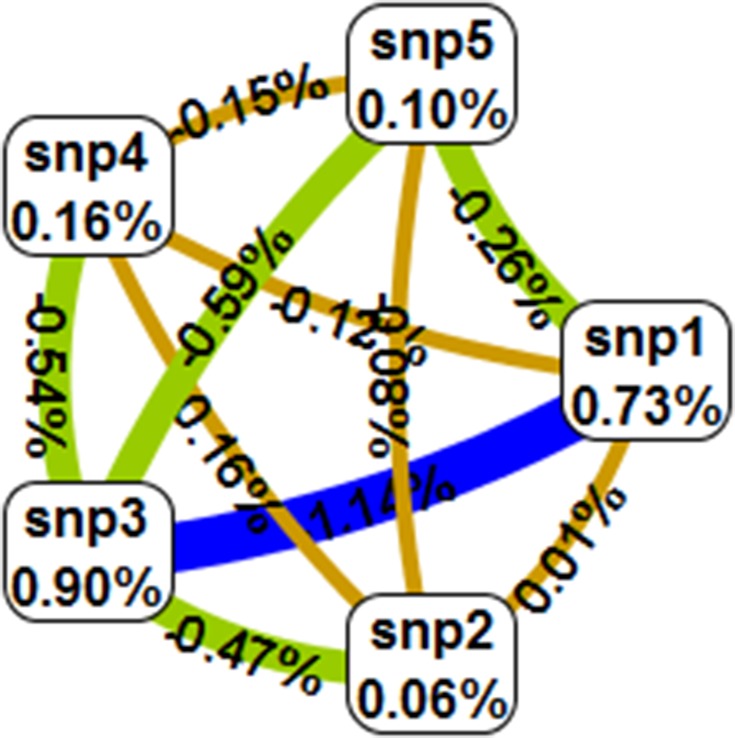
Table 9 summarises the MDR analysis evaluating the results of all possible combinations of the studied polymorphisms. According to MDR analysis, the best MDR model included all the five studied polymorphisms XRCC1 (p.Arg399Gln, p.Arg194Trp, p.Arg280His), XRCC2 (p.Arg188His) and XRCC3 (p.Thr241Met). The model had a testing balance accuracy of 0.4648 and a training balance accuracy of 0.5905 with a maximum cross-validation consistency of 10/10. However, the model was not statistically significant (p=0.9). Figure 2 depicts the interaction map of all genes, based on entropy measures between individual genes showing a redundancy effect.
Table 9.
Multifactor Dimensionality Reduction (MDR) Analysis
| Locus Number | Model | Training Balance Accuracy | Testing Balance Accuracy | Cross Validation Consistency | p value* |
|---|---|---|---|---|---|
| 1 | SNP1 | 0.5367 | 0.4965 | 7/10 | 0.37 |
| 2 | SNP1-SNP3 | 0.5494 | 0.4538 | 4/10 | 0.99 |
| 3 | SNP1-SNP3-SNP5 | 0.5621 | 0.4547 | 5/10 | 0.98 |
| 4 | SNP1-SNP2-SNP3-SNP5 | 0.5764 | 0.4289: | 6/10 | 0.98 |
| 5 | SNP1-SNP2-SNP3-SNP4-SNP5 | 0.5905 | 0.4648 | 10/10 | 0.98 |
Notes: SNP1: XRCC1 p.Arg399Gln, SNP2: XRCC1 p.Arg194Trp, SNP3: XRCC1 p.Arg280His, SNP4: XRCC2 Arg188His, SNP5: XRCC2 p.Thr241Met. *p values were based on 1000 permutations.
Figure 2.
Gene–Gene interaction map for esophageal cancer risk: Values inside nodes indicate information gain (IG) of individual attributes or main effects, whereas values between nodes show IG of pairwise combinations of attributes or interaction effects. Positive entropy (red) indicates strong interaction (not found), (yellow) weak interaction while negative entropy (green) indicates redundancy and blue shows no interactions.
Notes: SNP1: XRCC1 p.Arg399Gln, SNP2: XRCC1 p.Arg194Trp, SNP3: XRCC1 p.Arg280His, SNP4: XRCC2 Arg188His, SNP5: XRCC2 p.Thr241Met.
Discussion
In India, the esophagus is a major cancer site (NCRP 2016) with a majority being Squamous Cell Carcinoma (SCC) and a comparatively less frequency of adenocarcinoma patients.32,33 Similarly, the predominant type of EC in our study was SCC (88%) with only 12% adenocarcinoma patients.
In India, the inter-individual differences in susceptibility to cancer due to the genetic polymorphisms in XRCC1 previously found.34 As DNA-repair gene polymorphisms play a very important role in carcinogenesis, we carried out this case-control study to evaluate whether XRCC1 (p.Arg399Gln, p.Arg194Trp and p.Arg280His), XRCC2 (p.Arg188His), and XRCC3 (p.Thr241Met) gene polymorphisms modulate the risk of esophageal cancer. The varied results reported world over for these polymorphisms has been attributed to variations in DNA-repair gene polymorphisms amongst different ethnicities.35–39 The population of Punjab, North India are an Indo-Aryan ethnic group with Caucasian, Indoscythian racial admixture. The main caste groups prevalent in Punjab are Banias (merchant and moneylender), Brahmins (traditionally priestly but now involved in various occupations), Jatt Sikhs, which constitute the largest part of Sikh community (mainly agriculturist), and Scheduled caste (various occupations).40 In the present study, about 60% of the subjects belonged to Jatt Sikh Caste group but the subgroup analysis based on castes was not done due to unequal distribution of the samples.
The XRCC1p.Arg399Gln polymorphism is involved in various protein–protein interactions41 and higher sister chromatid exchanges and DNA adducts.42 In the present study, we found that A allele and the AA genotype of XRCC1p.Arg399Gln polymorphism was associated with a decreased risk of esophageal cancer. Furthermore, a reduced risk was observed in the recessive model (GG+GAvsAA). When compared with previously reported studies from India, the results of our study were consistent with some studies, especially from North India but contrary to others (Table 1). Very few studies relating the XRCC1 Arg399Gln polymorphism with esophageal cancer risk are available from which only one is from India. Among previous reports from North India on EC; a study of Chandigarh region34 found association with a decreased risk of XRCC1 p.Arg399Gln, another study from Uttar Pradesh43 also reported Arg/Gln (p=0.03, OR= 0.62) and Gln/Gln (p=0.003, OR=0.37) genotype to be associated with a decreased risk of gall bladder cancer; a study from Kashmir on colorectal cancer44 also reported a protective role of AA genotype. Similar results were reported in three other studies from North India on head and neck cancer,45 hepatocellular carcinoma46 and lung cancer.47
Contrary to the results of the present study, some previous studies from North India have reported an increased risk with AA genotype of XRCC1p.Arg399Gln polymorphism in lung cancer,48 head and neck cancer,49 colorectal cancer50 and prostate cancer.51 Similarly, some studies from South India reported an association of increased risk with the AA genotype in gastric cancer,52 colorectal cancer,29 lung cancer,53,54 cervical cancer,55 Naso-pharyngeal cancer,56 acute lymphoblastic leukemia57 and acute myeloid leukemia.58 However, few studies from South India reported no association of XRCC1p.Arg399Gln polymorphism with any of cancer (Table 1).
Results of the present study on XRCC1 p.Arg399Gln polymorphism is in agreement with the studies from different parts of the globe like esophageal cancer in Han Chinese,59 colorectal adenocarcinoma in Norwegian population,60 gallbladder cancer,61 and non-melanoma skin cancers.62 In contrast, some previous studies have reported the association of Arg399Gln polymorphism with an increased risk of esophageal,63 stomach and oral cancers,64 colorectal cancers in Korean, Egyptian and Japanese populations,65–67 lung cancer68 and breast cancer.22 However, three studies did not find any association between p.Arg399Gln polymorphism and cancer of the esophagus,69 gall bladder70 and breast.71 We did not observe any significant association of XRCC1p.Arg194Trp polymorphism with EC risk in the present study. The results were similar to two previous Indian studies.52,72 Among international studies, no association of XRCC1 p.Arg194Trp has been reported with EC risk in the population of North Carolina,73 gastric cancer in Korean population74 and breast cancer in Caucasian women.75 On the contrary, Trp allele has been reported to be associated with an increased risk of gastric cancer in the Chinese population.76 XRCC1 is a protein involved in Base excision repair and efficient repair of DNA single-strand breaks. In the present study, we observed a lower risk of esophageal cancer associated with p.Arg399Gln polymorphism of XRCC1. A relationship between polymorphism in XRCC1Arg399Gln and increased rate of apoptosis has been reported in patients of ulcerative colitis77 and in schizophrenia patients.78 The alterations to the DNA repair system could not only suppress it to accomplish the repair process, but it would also become more vulnerable to carry apoptosis and help in wiping out the damaged cells. The increased rates of the apoptosis results into the elimination of potential premalignant cells and hence, the XRCC1 Gln399 may play a protective role in esophageal cancer risk.
The A allele of XRCC1p.Arg280His was associated with an increased risk of esophageal cancer in the present study. Reported studies in different populations and different cancers have contradictory results for this polymorphism. From the North Indian population, p.Arg280His has been associated with an increased risk of hepatocellular carcinoma46 but no association has been reported with SCC head and neck.45 From a South Indian population, no significant association with breast cancer risk was reported.22 A meta-analysis within the Asian population has reported an association of p.Arg280His polymorphism with bladder cancer risk.79 Another meta-analysis reported the heterozygote and the dominant model to be associated with prostate cancer risk.80 A allele was associated with an increased risk of adenomas in the Norwegian population.60 A study on the Chinese population reported no association between p.Arg280His polymorphism and ESCC.81
XRCC2, an essential protein in the HRR pathway is required for the formation of RAD51 focus, with involvement in tumor progression.13 No association of XRCC2 p.Arg188His with EC has been observed in the present study, which is the first report from India for esophageal cancer risk. In previous reports from India, no association of XRCC2 p.Arg188His polymorphism was reported with nasopharyngeal carcinoma47 whereas another study found an association of GA genotype with increased risk of SCC head and neck.49 The contradictory reports from different parts of the world have shown that XRCC2 p.Arg188His polymorphism was associated with a significantly increased risk of pharyngeal cancer16 and breast cancer12 but not with bladder cancer,82 colorectal adenoma,83 skin cancer,84 thyroid cancer85,86 and breast cancer.87 Contrary to this, a protective role of GA and AA genotype in Caucasian breast cancer females of Cyprus has been reported.88
XRCC3 gene mainly repairs using the HR pathway and in vitro studies revealed high sensitivity to DNA damaging agents in cells with XRCC3 gene knockouts.89 No association of XRCC3 p.Thr241Met polymorphism with esophageal cancer has been observed in the present study. A previous study from the north-eastern region of India has reported no association of this polymorphism with breast cancer.90 From Indian subcontinent, two populations practicing consanguineous marriages reported an association with increased risk, for colorectal cancer in Kashmiri population50 and breast cancer in Pakistani population.91 For XRCC3 p.Thr241Met polymorphism, no association was reported with gastric cancer in Italian population92 and with colorectal cancer in the West Algerian population93 but an association with an increased risk was reported for lung cancer in an Italian population,23 oral SCC in Thai population,94 and gastric cancer in the Chinese population.95 On the other hand, a protective role of CT and TT genotype has been reported in colorectal cancer in Polish population96 and Basal cell carcinoma of the skin in population from Hungary, Romania and Slovakia.97
The haplotype analysis in the present study shows an association of haplotype GGT of XRCC1 gene (Arg399-Arg280-194Trp of p.Arg399Gln, p.Arg280His, and p.Arg194Trp polymorphisms) with a risk of EC, however, the association was not statistically significant (Table 7). Among previous reports from India, a study from Andhra Pradesh (South India) also found no association of any haplotype with Chronic Myeloid Leukemia.98 However, a study from West Bengal (East India) reported CAG haplotype (Arg194-His280-Arg399) to have reduced risk against gastric cancer.52 A study from Uttar Pradesh (North India) reported CGA and CAG haplotype of XRCC1 (Arg194-His280-Arg399) to be associated with prostate cancer and haplotype CGA to be associated with bladder cancer.38
Post radiotherapy in a cancer patient, radiation injury may develop months to years later after treatment, with numerous individual manifestations. The XRCC1 codon 399 Arg/Arg genotype has been associated with increased risk of acute radiation dermatitis in nasopharyngeal carcinoma patients treated with intensity-modulated radiation therapy.99 Hence a patient with the variant AA genotype of XRCC1p.Arg399Gln polymorphism might be at lower risk for radiation injury. The present study indicates a protective role of the XRCC1 p.Arg399Gln towards the development of EC. Therefore, XRCC gene polymorphisms can also be predicting factor in therapeutic decisions for individualized therapy in EC. Some of the results are at variance from reporting studies in Indian Sub-continent and other Asian countries. These might be due to the ethnic variations in the population residing in Punjab, Northwest India, other states of India and the rest of Asia. Inhabitants of Punjab and Northwestern states of India have an admixture of Indo-Scythian and Caucasian racial elements with more similarity with the central Asian populations as compared to populations in Middle India, South India or eastern parts of India.25,100
Conclusion
The present study being the first report from India, providing baseline data about five genetic polymorphisms in three DNA repair genes XRCC1, XRCC2 and XRCC3 modulating overall esophageal cancer susceptibility in ethnic Punjabi Indian subjects. The future utility of this data in screening the population for EC risk is the major strength of the study. The results have also been adjusted for various confounding factors. The limitation is a lack of stratification of subjects on the basis of endogamous caste groups of Punjab due to small cross-sectional sample in individual caste group. A possible interaction between XRCC polymorphisms and environmental factors on cancer risk was investigated in several studies. It is of great interest to evaluate gene–environment interactions to examine the exact roles of genetic polymorphisms in cancer. However, lacking the original data limited our further evaluation of potential gene–environment interactions. The results suggest a role of XRCC gene polymorphisms in esophageal cancer risk and a need to confirm our findings with higher sample size in different ethnic groups inhabiting different geographical areas of India.
Acknowledgments
We are grateful to the participants of the study. The research funding from UGC-UPE and UGC- SAP DRS II to Vasudha Sambyal and Kamlesh Guleria and research fellowship to Jagjeet Kaur is highly acknowledged.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):359–386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Samarasam I. Esophageal cancer in India, current status and future perspectives. Int J Adv Med Health Res. 2017;4:5–10. doi: 10.4103/IJAMR.IJAMR_19_17 [DOI] [Google Scholar]
- 3.Murtaza I, Mushtaq D, Margoob MA, et al. A study on p53 gene alterations in esophageal squamous cell carcinoma and their correlation to common dietary risk factors among population of the Kashmir valley. World J Gastroenterol. 2006;12:4033–4037. doi: 10.3748/wjg.v12.i25.4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154 [DOI] [PubMed] [Google Scholar]
- 5.London RE. The structural basis of XRCC1-mediated DNA repair. DNA Repair (Amst). 2015;30:90–103. doi: 10.1016/j.dnarep.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen MR, Zdzienicka MZ, Mohrenweiser H, Thompson LH, Thelen MP. Mutations in hamster single-strand break repair gene XRCC1 causing defective DNA repair. Nucleic Acids Res. 1998;26:1032–1037. doi: 10.1093/nar/26.4.1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pramanik S, Devi S, Chowdhary S, Surendran S, Krishnamurthi K, Chakrabarti T. DNA repair gene polymorphisms at XRCC1, XRCC3, XPD and OGG1 loci in Maharashtrian population of central India. Chemosphere. 2011;82:941–946. doi: 10.1016/j.chemosphere.2010.10.100 [DOI] [PubMed] [Google Scholar]
- 8.Moullan N, Cox DG, Angele S, Romestaing P, Gerard JP, Hall J. Polymorphisms in the DNA repair gene XRCC1, breast cancer risk and response to radiotherapy. Cancer Epidemiol Biomarkers Prev. 2003;12:1168–1174. [PubMed] [Google Scholar]
- 9.Ratnasinghe D, Yao SX, Tangrea JA, et al. Polymorphisms of the DNA repair gene XRCC1 and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10:119–123. [PubMed] [Google Scholar]
- 10.Parine NR, Pathan AA, Bobbarala V, Abduljaleel Z, Khan W, Alanazi M. DNA repair gene polymorphisms at XRCC1, XRCC3, XPD, and OGG1 Loci in the hyderabad population of India. Asian Pac J Cancer Prev. 2012;13:6469–6474. [DOI] [PubMed] [Google Scholar]
- 11.Braybrooke JP, Spink KG, Thacker J, Hickson ID. The RAD51 family member, RAD51L3, is a DNA-stimulated ATPase that forms a complex with XRCC2. J Biol Chem. 2000;275:29100–29106. doi: 10.1074/jbc.M002075200 [DOI] [PubMed] [Google Scholar]
- 12.Kuschel B, Auranen A, McBride S, et al. Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum Mol Genet. 2002;11:1399–1407. doi: 10.1093/hmg/11.12.1399 [DOI] [PubMed] [Google Scholar]
- 13.Jiao L, Hassan MM, Bondy ML, et al. XRCC2 and XRCC3 gene polymorphism and risk of pancreatic cancer. Am J Gastroenterol. 2008;103:360–367. doi: 10.1111/ajg.2008.103.issue-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auranen A, Song H, Waterfall C, et al. Polymorphisms in DNA repair genes and epithelial ovarian cancer risk. Int J Cancer. 2005;117(4):611–618. doi: 10.1002/(ISSN)1097-0215 [DOI] [PubMed] [Google Scholar]
- 15.Yen CY, Liu SY, Chen CH, et al. Combinational polymorphisms of four DNA repair genes XRCC1, XRCC2, XRCC3 and XRCC4 and their association with oral cancer in Taiwan. J Oral Pathol Med. 2008;37:271–277. doi: 10.1111/j.1600-0714.2007.00608.x [DOI] [PubMed] [Google Scholar]
- 16.Benhamou S, Tuimala J, Bouchardy C, Dayer P, Sarasin A, Hirvonen A. DNA repair gene XRCC2 and XRCC3 polymorphisms and susceptibility to cancers of the upper aerodigestive tract. Int J Cancer. 2004;112(5):901–904. doi: 10.1002/ijc.20474 [DOI] [PubMed] [Google Scholar]
- 17.Talar-wojnarowska R, Gąsiorowska A, Olakowski A, et al. Analysis of XRCC2 and XRCC3 gene polymorphisms in pancreatic cancer. Biomed Rep. 2016;4:236–240. doi: 10.3892/br.2015.550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Deng X, Wang Z, Wang Q, Liu Y. Genetic polymorphisms of DNA repair genes XRCC1 and XRCC3 and risk of colorectal cancer in Chinese population. Asian Pac J Cancer Prev. 2012;13:665–669. doi: 10.7314/APJCP.2012.13.2.665 [DOI] [PubMed] [Google Scholar]
- 19.Abdel-fatah T, Arora A, Gorguc I, et al. Are DNA repair factors promising biomarkers for personalized therapy in gastric cancer? Antioxid Redox Signal. 2013;18(18):2392–2398. doi: 10.1089/ars.2012.4873 [DOI] [PubMed] [Google Scholar]
- 20.Mao CF, Qian WY, Wu JZ, Sun DW, Tang JH. Association between the XRCC3 Thr241Met polymorphism and breast cancer risk: an updated meta-analysis of 36 case-control studies. Asian Pac J Cancer Prev. 2014;15(16):6613–6618. doi: 10.7314/APJCP.2014.15.16.6613 [DOI] [PubMed] [Google Scholar]
- 21.Liu HX, Li J, Ye BG. Correlation between gene polymorphisms of CYP1A1, GSTP1, ERCC2, XRCC1, and XRCC3 and susceptibility to lung cancer. Genet Mol Res. 2016;15(4). doi: 10.4238/gmr15048813 [DOI] [PubMed] [Google Scholar]
- 22.Chacko P, Rajan B, Joseph T, Methew BS, Pillai MR. Polymorphisms in DNA repair gene XRCC1 and increased genetic susceptibility to breast cancer. Breast Cancer Res Treat. 2005;89:15–21. doi: 10.1007/s10549-004-1004-x [DOI] [PubMed] [Google Scholar]
- 23.Improta G, Sgambato A, Bianchino G, et al. Polymorphisms of the DNA repair genes XRCC1 and XRCC3 and risk of lung and colorectal cancer, a case-control study in a Southern Italian population. Anticancer Res. 2008;28:291–294. [PubMed] [Google Scholar]
- 24.Mandarapu R, Konathala G. Base excision repair pathway gene polymorphisms in South Indian population. Int J Contemp Med Res. 2016;3:77–83. [Google Scholar]
- 25.Basu A, Mukherjee N, Roy S, et al. Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res. 2003;13(10):2277–2290. doi: 10.1101/gr.1413403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asthana S, Patil RS, Labani S. Tobacco-related cancers in India: A review of incidence reported from population-based cancer registries. Indian J Med Paediatr Oncol. 2016;37(3):152–157. doi: 10.4103/0971-5851.190357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bal MS, Bodal VK, Kaur J, Kaur M, Sharma S. Patterns of Cancer, A Study of 500 punjabi patients. Asian Pac J Cancer Prev. 2016;16:5107–5110. doi: 10.7314/APJCP.2015.16.12.5107 [DOI] [PubMed] [Google Scholar]
- 28.Adeli K, Ogbonna G. Rapid purification of human DNA from whole blood for potential application in clinical chemistry laboratories. Clin Chem. 1990;36(2):261–264. doi: 10.1093/clinchem/36.2.261 [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Zhao Y, Jiang J. Polymorphisms in the DNA repair genes XRCC1, XRCC3 and XPD, and colorectal cancer risk, a case–control study in an {I}ndian population. J Cancer Res Clin Oncol. 2010;136:1517–1525. doi: 10.1007/s00432-010-0809-8 [DOI] [PubMed] [Google Scholar]
- 30.Gok I, Baday M, Cetinkunar S, Kilic K, Bilgin BC. Polymorphisms in DNA repair genes XRCC2 and XRCC3 risk of gastric cancer in Turkey. Bosn J Basic Med Sci. 2014;14(4):214–218. doi: 10.17305/bjbms.2014.4.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhasin MK. Morphology to molecular anthropology: castes and tribes of India. Int J Hum Genet. 2009;9(3–4):145–230. doi: 10.1080/09723757.2009.11886070 [DOI] [Google Scholar]
- 32.Alsop BR, Sharma P. Esophageal cancer. Gastroenterol Clin North Am. 2016;45(3):399–412. doi: 10.1016/j.gtc.2016.04.001 [DOI] [PubMed] [Google Scholar]
- 33.Kolkman P, Stack A, McCarthy M, et al. Variations in the histopathologic type of esophageal carcinoma between the United States of America and India. Indian J Surg Oncol. 2016;7:52–55. doi: 10.1007/s13193-015-0468-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobti RC, Singh J, Kaur P, Pachouri SS, Siddiqui EA, Bindra HS. XRCC1 codon 399 and ERCC2 codon 751 polymorphism, smoking, and drinking and risk of esophageal squamous cell carcinoma in a North Indian population. Cancer Genet Cytogenet. 2007;175:91–97. doi: 10.1016/j.cancergencyto.2007.01.001 [DOI] [PubMed] [Google Scholar]
- 35.Duarte MC, Jucimara C, Regina A, Rossit B, Silva AE. Polymorphisms of the DNA repair genes XRCC1 and XRCC3 in a Brazilian population. Genet Mol. 2005;28:397–401. [Google Scholar]
- 36.Vettriselvi V, Vijayalakshmi K, Solomon PFD, Venkatachalam P. XRCC1 and XPD gene polymorphisms in a South Indian population. Asian Pac J Cancer Prev. 2007;8:283–286. [PubMed] [Google Scholar]
- 37.Gangwar R, Manchanda PK, Mittal RD. Implications of XRCC1, XPD and APE1 gene polymorphism in North Indian population, a comparative approach in different ethnic groups worldwide. Genetica. 2008;136:163–169. doi: 10.1007/s10709-008-9329-8 [DOI] [PubMed] [Google Scholar]
- 38.Mittal RD, Mandal RK, Gangwar R. Base excision repair pathway genes polymorphism in prostate and bladder cancer risk in North Indian population. Mech Ageing Dev. 2012;133:127–132. doi: 10.1016/j.mad.2011.10.002 [DOI] [PubMed] [Google Scholar]
- 39.Rao SK, Paul A, Sudarsan A, et al. Allele and genotype distributions of DNA repair gene polymorphisms in South Indian healthy population. Biomark Cancer. 2014;6:29–35. doi: 10.4137/BIC.S19681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sekhon IS. Punjabis: The People, Their History, Culture and Enterprise. New Delhi: Cosmo Publisher; 2000. [Google Scholar]
- 41.Shen M, Hung RJ, Brennan P, et al. Polymorphisms of the DNA repair genes XRCC1, XRCC3, XPD, interaction with environmental exposures, and bladder cancer risk in a case–control study in northern Italy. Cancer Epidemiol Biomarkers Prev. 2003;12:1234–1240. [PubMed] [Google Scholar]
- 42.Duell EJ, Wiencke JK, Cheng TJ, et al. Polymorphisms in the DNA repair genes XRCC1 and ERCC2 and biomarkers of DNA damage in human blood mononuclear cell. Carcinogenesis. 2000;21:965–971. doi: 10.1093/carcin/21.5.965 [DOI] [PubMed] [Google Scholar]
- 43.Srivastava A, Srivastava K, Pandey SN, Choudhuri G, Mittal B. Single-nucleotide polymorphisms of DNA repair genes OGG1 and XRCC1, association with gall bladder cancer in North Indian population. Ann Surg Oncol. 2009;16:1695–1703. doi: 10.1245/s10434-009-0354-3 [DOI] [PubMed] [Google Scholar]
- 44.Khan NP, Pandith AA, Yousuf A, et al. The XRCC1 Arg399Gln gene polymorphism and risk of colorectal cancer, a study in Kashmir. Asian Pac J Cancer Prev. 2013;14:6779–6782. doi: 10.7314/APJCP.2013.14.11.6779 [DOI] [PubMed] [Google Scholar]
- 45.Kumar A, Pant MC, Singh HS, Khandelwal S. Associated risk of XRCC1 and XPD cross talk and life style factors in progression of head and neck cancer in north Indian population. Mut Res. 2012;729:24–34. doi: 10.1016/j.mrfmmm.2011.09.001 [DOI] [PubMed] [Google Scholar]
- 46.Kiran M, Saxena R, Chawla YK, Kaur J. Polymorphism of DNA repair gene XRCC1 and hepatitis-related hepatocellular carcinoma risk in Indian population. Mol Cell Biochem. 2009;327:7–13. doi: 10.1007/s11010-009-0035-3 [DOI] [PubMed] [Google Scholar]
- 47.Singh A, Singh N, Behera D, Sharma S. Association and multiple interaction analysis among five XRCC1 polymorphic variants in modulating lung cancer risk in North Indian population. DNA Repair (Amst). 2016;47:30–41. doi: 10.1016/j.dnarep.2016.09.006 [DOI] [PubMed] [Google Scholar]
- 48.Uppal V, Mehndiratta M, Mohapatra D, Grover RK, Puri D. XRCC-1 gene polymorphism (Arg399Gln) and susceptibility to development of lung cancer in cohort of North Indian population A pilot study. J Clin Diagn Res. 2014;8:17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choudhury JH, Choudhury B, Kundu S, Ghosh SK. Combined effect of tobacco and DNA repair genes polymorphisms of XRCC1 and XRCC2 influence high risk of head and neck squamous cell carcinoma in northeast Indian population. Med Oncol. 2014;31(8):67. doi: 10.1007/s12032-014-0067-8 [DOI] [PubMed] [Google Scholar]
- 50.Nissar S, Lone TA, Banday MZ, et al. Arg399Gln polymorphism of XRCC1 gene and risk of colorectal cancer in Kashmir, A case control study. Oncol Lett. 2013;5:959–963. doi: 10.3892/ol.2013.1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berhane N, Sobti RC, Mahdi SA. DNA repair genes polymorphism (XPG and XRCC1) and association of prostate cancer in a north Indian population. Mol Biol Rep. 2012;39(3):2471–2479. doi: 10.1007/s11033-011-0998-5 [DOI] [PubMed] [Google Scholar]
- 52.Ghosh S, Ghosh S, Bankura B, et al. Association of DNA repair and xenobiotic pathway gene polymorphisms with genetic susceptibility to gastric cancer patients in West Bengal India. Tumour Biol. 2016;37:9139–9149. doi: 10.1007/s13277-015-4780-5 [DOI] [PubMed] [Google Scholar]
- 53.Sreeja L, Syamala VS, Syamala V, et al. Prognostic importances of DNA repair gene polymorphisms of XRCC1 Arg399Gln and XPD Lys751Gln in lung cancer patients from India. J Cancer Res Clin Oncol. 2008;134:645–652. doi: 10.1007/s00432-007-0328-4 [DOI] [PubMed] [Google Scholar]
- 54.Natukula K, Jamil K, Pingali UR, Attili VS, Madireddy UR. The codon 399 Arg/Gln XRCC1 polymorphism is associated with lung cancer in Indians. Asian Pac J Cancer Prev. 2013;14:5275–5279. doi: 10.7314/APJCP.2013.14.9.5275 [DOI] [PubMed] [Google Scholar]
- 55.Konathala G, Mandarapu R, Godi S. Data on polymorphism of XRCC1 and cervical cancer risk from South India. Data Brief. 2017;10:11–13. doi: 10.1016/j.dib.2016.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh SR, Ghosh SK. Polymorphisms of XRCC1 and XRCC2 DNA repair genes and interaction with environmental factors influence the risk of Nasopharyngeal carcinoma in Northeast India. Asian Pac J Cancer Prev. 2016;17:2811–2819. [PubMed] [Google Scholar]
- 57.Joseph T, Kusumakumary P, Chacko P, Abraham A, Pillai MR. DNA repair gene XRCC1 polymorphisms in childhood acute lymphoblastic leukemia. Cancer Lett. 2005;217:17–24. doi: 10.1016/j.canlet.2004.06.055 [DOI] [PubMed] [Google Scholar]
- 58.Santhi S, Sangeetha V, Sureshkumar R, et al. Risk effects of XRCC1 Arg399Gln and XPD Lys751Gln gene polymorphisms in de novo acute myeloid leukemia - A study from India. Indian J Biotechnol. 2017;16:275–283. [Google Scholar]
- 59.Xing D, Qi J, Miao X, Lu W, Tan W, Lin D. Polymorphisms of DNA repair genes XRCC1 and XPD and their associations with risk of esophageal squamous cell carcinoma in a Chinese population. Int J Cancer. 2002;100:600–605. doi: 10.1002/ijc.10528 [DOI] [PubMed] [Google Scholar]
- 60.Skjelbred CF, Saebø M, Wallin H, et al. Polymorphisms of the XRCC1, XRCC3 and XPD genes and risk of colorectal adenoma and carcinoma in a Norwegian cohort, a case-control study. BMC Cancer. 2006;6:67–76. doi: 10.1186/1471-2407-6-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stern MC, Umbach DM, van Gils CH, Lunn RM, Taylor JA. DNA repair gene XRCC1 polymorphisms, smoking and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10:125–132. [PubMed] [Google Scholar]
- 62.Nelson HH, Kelsey KT, Mott LA, Karagas MR. The XRCC1 polymorphism, sunburn, and non-melanoma skin cancer, evidence of gene-environment interaction. Cancer Res. 2002;62:152–160. [PubMed] [Google Scholar]
- 63.Li S, Deng Y, You JP, et al. XRCC1 Arg399Gln, Arg194Trp, and Arg280His polymorphisms in esophageal cancer risk, a meta-analysis. Digest Dis Sci. 2013;58:1880–1890. [DOI] [PubMed] [Google Scholar]
- 64.Shu XO, Cai Q, Gao YT, Wen W, Jin F, Zheng W. A population based case control study of the Arg399Gln polymorphism in DNA repair gene XRCC1 and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2003;12:1462–1467. [PubMed] [Google Scholar]
- 65.Abdel-rahman SZ, Soliman AS, Bondy ML, et al. Inheritance of the 194Trp and the 399Gln variant alleles of the DNA repair gene XRCC1 are associated with increased risk of early-onset colorectal carcinoma in Egypt. Cancer Lett. 2000;159(1):79–86. doi: 10.1016/S0304-3835(00)00537-1 [DOI] [PubMed] [Google Scholar]
- 66.Hong YC, Lee H, Kim WC, Choi SK, ZHa W, Shin SK. Polymorphisms of XRCC1 gene, alcohol consumption and colorectal cancer. Int J Cancer. 2005;116:428–432. doi: 10.1002/ijc.21019 [DOI] [PubMed] [Google Scholar]
- 67.Yin G, Morita M, Ohnaka K, et al. Genetic polymorphisms of XRCC1, alcohol consumption, and the risk of colorectal cancer in Japan. J Epidemiol. 2011;22:64–71. doi: 10.2188/jea.JE20110059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park JY, Lee SY, Jeon HS, et al. Polymorphism of the DNA repair gene XRCC1 and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2002;11:23–27. [PubMed] [Google Scholar]
- 69.Yu H-P, Zhang X-Y, Wang X-L, et al. DNA repair gene XRCC1 polymorphisms, smoking and esophageal cancer risk. Cancer Detect Prev. 2004;28:194–199. doi: 10.1016/j.cdp.2004.01.004 [DOI] [PubMed] [Google Scholar]
- 70.Huang WY, Gao YT, Rashid A, et al. Selected base excision repair gene polymorphisms and susceptibility to biliary tract cancer and biliary stones, a population-based case-control study in China. Carcinogenesis. 2008;29:100–105. doi: 10.1093/carcin/bgm247 [DOI] [PubMed] [Google Scholar]
- 71.Karolina PS, Stanczyk M, Kusinska R, Kordek R, Majsterek I. Association of the Arg194Trp and the Arg399Gln polymorphisms of the XRCC1 gene with risk occurrence and the response to adjuvant therapy among Polish women with breast cancer. Clin Breast Cancer. 2013;13:61–68. doi: 10.1016/j.clbc.2012.09.019 [DOI] [PubMed] [Google Scholar]
- 72.Pavithra D, Gautam M, Rama R, et al. TGFβ C-509T, TGFβ T869C, XRCC1 Arg194Trp, IKBα C642T, IL4 C-590T Genetic polymorphisms combined with socio-economic, lifestyle, diet factors and gastric cancer risk: A case control study in South Indian population. Cancer Epidemiol. 2018;53:21–26. doi: 10.1016/j.canep.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 73.Lunn RM, Langlois RG, Hsieh LL, Thompson CL, Bell DA. XRCC1 polymorphisms, effects on aflatoxin B-DNA adducts and glycophorin A variant frequency. Cancer Res. 1999;59:2557–2561. [PubMed] [Google Scholar]
- 74.Jin E-H, Kim J, Lee S-I, Hong JH. Association between polymorphisms in APE1 and XRCC1 and the risk of gastric cancer in Korean population. Int J Clin Exp Med. 2015;8:11484–11489. [PMC free article] [PubMed] [Google Scholar]
- 75.Sterpone S, Mastellone V, Padua L, et al. Single-nucleotide polymorphisms in BER and HRR genes, XRCC1 haplotypes and breast cancer risk in Caucasian women. J Cancer Res Clin Oncol. 2010;136:631–636. doi: 10.1007/s00432-010-0791-1 [DOI] [PubMed] [Google Scholar]
- 76.Wen -Y-Y, Pan X-F, Loh M, et al. ADPRT Val762Ala and XRCC1 Arg194Trp Polymorphisms and risk of gastric cancer in Sichuan of China. Asian Pac J Cancer Prev. 2012;13:2139–2144. doi: 10.7314/APJCP.2012.13.5.2139 [DOI] [PubMed] [Google Scholar]
- 77.Bardia A, Tiwari SK, Gunisetty S, et al. Functional polymorphisms in XRCC-1 and APE-1 contribute to increased apoptosis and risk of ulcerative colitis. Inflamm Res. 2012;61(4):359–365. doi: 10.1007/s00011-011-0418-2 [DOI] [PubMed] [Google Scholar]
- 78.Catts VS, Catts SV, McGrath JJ, et al. Apoptosis and schizophrenia: a pilot study based on dermal fibroblast cell lines. Schizophr Res. 2006;84(1):20–28. doi: 10.1016/j.schres.2006.03.016 [DOI] [PubMed] [Google Scholar]
- 79.Fang Z, Chen F, Wang X, Yi S, Chen W, Ye G. XRCC1 Arg194Trp and Arg280His polymorphisms increase bladder cancer risk in Asian population, evidence from a meta-analysis. PLoS One. 2013;8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yan J, Wang X, Tao H, Deng Z, Yang W, Lin F. Meta-analysis of the relationship between XRCC1-Arg399Gln and Arg280His polymorphisms and the risk of prostate cancer. Sci Rep. 2015;5:9905. doi: 10.1038/srep09905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhai XD, Mo YN, Xue XQ, et al. XRCC1 codon 280 and ERCC2 codon 751 polymorphisms and risk of esophageal squamous cell carcinoma in a Chinese population. Bull Cancer. 2009;96:61–65. doi: 10.1684/bdc.2009.0952 [DOI] [PubMed] [Google Scholar]
- 82.Matullo G, Guarrera S, Sacerdote C, et al. Polymorphisms/haplotypes in DNA repair genes and smoking, a bladder cancer case–control study. Cancer Epidemiol Biomarkers Prev. 2005;14:2569–2578. doi: 10.1158/1055-9965.EPI-05-0189 [DOI] [PubMed] [Google Scholar]
- 83.Tranah GJ, Giovannucci E, Ma J, Fuchs C, Hankinson SE, Hunter DJ. XRCC2 and XRCC3 polymorphisms are not associated with risk of colorectal adenoma. Cancer Epidemiol Biomarkers Prev. 2004;13:1090LP- 1091. [PubMed] [Google Scholar]
- 84.Han J, Hankinson SE, Hunter DJ, De Vivo I. Genetic variations in XRCC2 and XRCC3 are not associated with endometrial cancer risk. Cancer Epidemiol Biomarkers Prev. 2004;13:330–331. doi: 10.1158/1055-9965.EPI-03-0332 [DOI] [PubMed] [Google Scholar]
- 85.Bastos HN, Antao MR, Silva SN, et al. Association of polymorphisms in genes of the homologous recombination DNA repair pathway and thyroid cancer risk. Thyroid. 2009;19(10):1067–1075. doi: 10.1089/thy.2009.0099 [DOI] [PubMed] [Google Scholar]
- 86.Yan L, Li Q, Li X, Ji H, Zhang L. Association studies between XRCC1, XRCC2, XRCC3 polymorphisms and differentiated thyroid carcinoma. Cell Physiol Biochem. 2016;38:1075–1084. doi: 10.1159/000443058 [DOI] [PubMed] [Google Scholar]
- 87.Yu KD, Chen AX, Qiu LX, Fan L, Yang C, Shao ZM. XRCC2 Arg188His polymorphism is not directly associated with breast cancer risk, evidence from 37,369 subjects. Breast Cancer Res Treat. 2010;123:219–225. doi: 10.1007/s10549-010-0753-y [DOI] [PubMed] [Google Scholar]
- 88.Loizidou MA, Michael T, Neuhausen S, et al. Genetic polymorphisms in the DNA repair genes XRCC1, XRCC2 and XRCC3 and risk of breast cancer in Cyprus. Breast Cancer Res Treat. 2008;112:575–579. doi: 10.1007/s10549-007-9881-4 [DOI] [PubMed] [Google Scholar]
- 89.Singleton BK, Griffin CS, Thacker J. Clustered DNA damage leads to complex genetic changes in irradiated human cells. Cancer Res. 2002;62(21):6263–6269. [PubMed] [Google Scholar]
- 90.Devi KR, Ahmed J, Narain K, et al. DNA repair mechanism gene, XRCC1A (Arg194Trp) but not XRCC3 (Thr241Met) polymorphism increased the risk of breast cancer in premenopausal females A case-control study in Northeastern region of India. Technol Cancer Res Treat. 2017;16:1150–1159. doi: 10.1177/1533034617736162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qureshi Z, Mahjabeen I, Baig R, Kayani M. Correlation between Selected XRCC2, XRCC3 and RAD51 gene polymorphisms and primary breast cancer in women in Pakistan. Asian Pac J Cancer Prev. 2014;15:10225–10229. doi: 10.7314/APJCP.2014.15.23.10225 [DOI] [PubMed] [Google Scholar]
- 92.Palli D, Polidoro S, D’errico M, et al. Polymorphic DNA repair and metabolic genes, a multigenic study on gastric cancer. Mutagenesis. 2010;25:569–575. doi: 10.1093/mutage/geq042 [DOI] [PubMed] [Google Scholar]
- 93.Moghtit FZ, Aberkane MS, Morvan L, et al. No association between XRCC3 Thr241Met and XPD Lys751Gln polymorphisms and the risk of colorectal cancer in west Algerian population, a case-control study. Medical Oncol. 2014;31:942–949. doi: 10.1007/s12032-014-0942-3 [DOI] [PubMed] [Google Scholar]
- 94.Kietthubthew S, Sriplung H, Au WW, Ishida T. Polymorphism in DNA repair genes and oral squamous cell carcinoma in Thailand. Int J Hyg Environ Health. 2006;209:21–29. doi: 10.1016/j.ijheh.2005.06.002 [DOI] [PubMed] [Google Scholar]
- 95.Huang GP, Zheng ZL, Cai L. DNA repair gene XRCC3 Thr241Met polymorphism and susceptibility to cardia and non-cardia gastric cancer, a case-control study. Zhonghua Liu Xing Bing XueZaZhi. 2006;27:420–423. [PubMed] [Google Scholar]
- 96.Mucha B, Przybylowska-sygut K, Dziki AJ, Dziki LS, Majsterek I. Association of Thr241Met polymorphism of XRCC3 gene with risk of colorectal cancer in the polish population. Pol J Pathol. 2013;64:185–190. doi: 10.5114/pjp.2013.38137 [DOI] [PubMed] [Google Scholar]
- 97.Thiumaran RK, Bermejo JL, Rudnai P, et al. Single nucleotide polymorphisms in DNA repair genes and basal cell carcinoma of skin. Carcinogenesis. 2006;27:1676–1681. doi: 10.1093/carcin/bgi381 [DOI] [PubMed] [Google Scholar]
- 98.Annamaneni S, Gorre M, Kagita S, et al. Association of XRCC1 gene polymorphisms with chronic myeloid leukemia in the population of Andhra Pradesh, India. Hematology. 2013;18(3):163–168. doi: 10.1179/1607845412Y.0000000040 [DOI] [PubMed] [Google Scholar]
- 99.Chen H, Wu M, Li G, Hua L, Chen S, Huang H. Association between XRCC1 single-nucleotide polymorphism and acute radiation reaction in patients with nasopharyngeal carcinoma. Medicine. 2017;96:e8202. doi: 10.1097/MD.0000000000008202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Majumder P. Ethnic populations of India as seen from an evolutionary perspective. J Biosci. 2001;26:533–545. doi: 10.1007/BF02704750 [DOI] [PubMed] [Google Scholar]
- 101.Datkhile KD, Bhosale SJ, Patil MN, Khamkar TS, Durgawale PP, Kakade SV. Genetic polymorphism of DNA repair genes (XRCC1, XRCC2 & XRCC3) in breast cancer patients from rural Maharashtra. Int J Health Sci Res. 2017;7:281–290. [Google Scholar]
- 102.Nishank SS. Association of DNA damage repair gene polymorphisms hOGG1, XRCC1and p53 with sickle cell disease patients in India. Mediterr J Hematol Infect Dis. 2015;7:e2015046. doi: 10.4084/mjhid.2015.046 [DOI] [PMC free article] [PubMed] [Google Scholar]