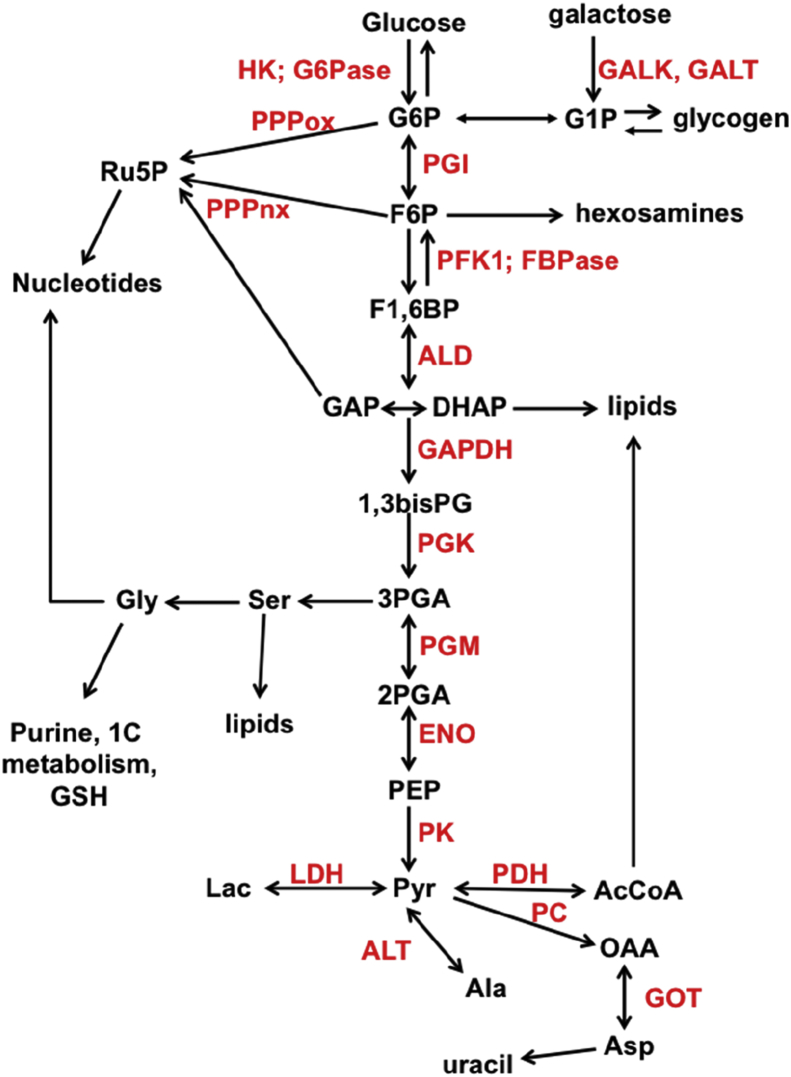
Figure 1.
Metabolic subnetwork centered on glucose metabolism. Double headed arrows represent reversible reactions catalyzed by the same enzyme; two arrows represent reactions catalyzed by different enzymes. Enzyme names are in red. G6P: glucose-6-phosphate; F6P fructose-6-phosphate; F1,6BP fructose-1,6-bisphosphate; GAP glyceraldehyde-3-phosphate; DHAP dihydroxyacetone phosphate; 1,3bisPG 1,3-bisphosphoglycerate; 2PGA 2-phosphogycerate; PEP phosphoenolpyruvate; Pyr pyruvate; OAA oxaloacetate; AcCoA acetyl CoA; Lac lactate; Ru5P ribose-5-phosphate; GSH reduced glutathione; GALK galactose 1 kinase; GALT galcaose-1-phosphate uridyltransferase; HK hexokinase; G6Pase glucose-6-phosphatase; PGI phosphoglucose isomerase; PFK1 phosphofructokinase 1; FBPase fructose 1,6 bisphosphatase; ALD aldolase; GAPDH glyceraldehyde-3- phosphate dehydrogenase; PGK phosphoglycerate kinase; PGM phosphoglycerate mutase; ENO enolase; PK pyruvate kinase; LDH lactate dehydrogenase; ALT alanine aminotransferase; PC pyruvate carboxylase; PDH pyruvate dehydrogenase; GOT glutamate oxaloacetate aminotransferase; PPPox oxidative branch of the pentose phosphate pathway; PPPnx non-oxidative branch of the pentose phosphate pathway.