Fig. 1. Nucleotide excision repair in Escherichia coli.
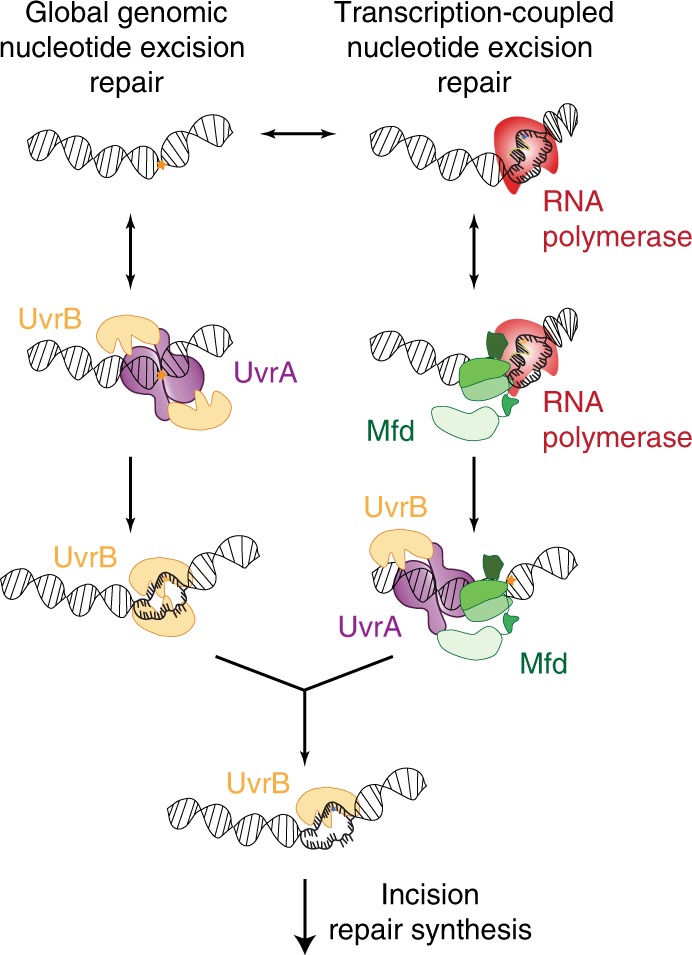
Damage detection in nucleotide excision repair in E. coli proceeds via global damage surveillance executed by UvrA2(B), and RNA polymerase (RNAP) transcribing damaged template DNA. The UvrA dimer loads UvrB, which verifies the presence of DNA damage in a strand-specific manner. Alternately, stalled elongation complexes at the site of DNA damage are rescued by the transcription-repair coupling factor Mfd, which in turn recruits UvrA2(B) to the site of the stalled RNAP. This is followed by strand-specific loading of UvrB at the site of the lesion. Following damage verification by UvrB, a single-stranded patch of DNA containing the damage is incised by the UvrC endonuclease. This is followed by repair synthesis and ligation coordinated by UvrD, PolI and LigA.
