Fig. 3. Measurements of Mfd-YPet binding lifetimes in cells expressing mutant UvrA deficient in ATP binding and hydrolysis.
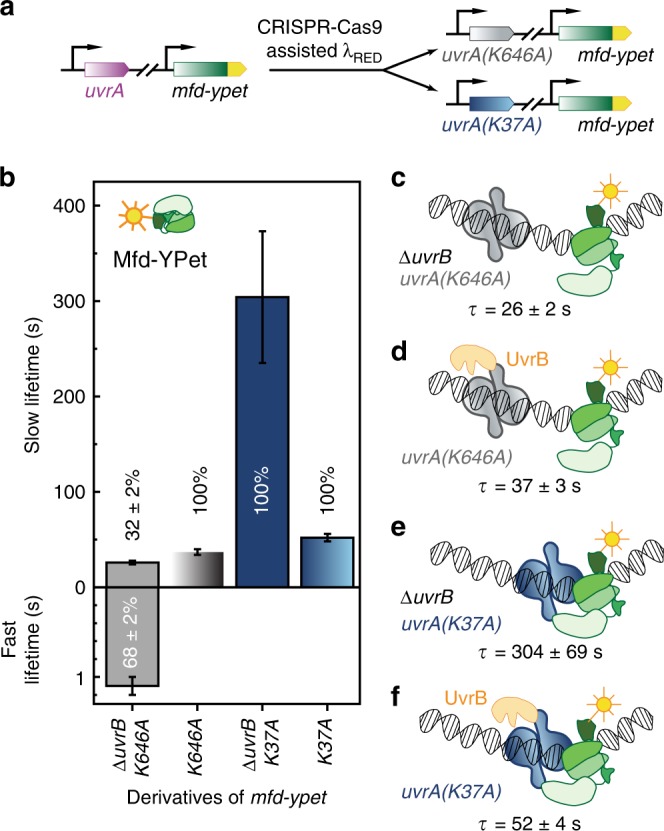
a The wild-type uvrA allele was replaced by the mutant allele in mfd-YPet cells, so that mutant cells either express the distal ATPase mutant UvrA(K646A) or the proximal ATPase mutant UvrA(K37A) from the chromosome. b Bar plots show lifetimes of DNA-bound Mfd-YPet in the corresponding genetic backgrounds. Where two kinetic sub-populations are detected, the fast lifetime is displayed in the lower panel. Numbers in percentages represent the amplitudes of kinetic sub-populations (see Supplementary Figs. 2, 3). c Lifetime of Mfd-YPet in mfd-YPet uvrA(K646A) ΔuvrB (26 ± 2 s; Nrepeats = 6 totaling nobs = 23,763 individual observations) is similar to that of Mfd-YPet in ΔuvrA cells (29 ± 2 s21), suggesting the distal ATPase mutant (gray) is unable to interact with Mfd (green). d Lifetime of Mfd-YPet in the presence of UvrA(K646A) and UvrB (orange) (37 ± 3 s; Nrepeats = 11 totaling nobs = 36,073 individual observations) also resembles that of Mfd-YPet in ΔuvrA cells. e Mfd-YPet is arrested in the presence of the proximal ATPase mutant UvrA(K37A) in cells lacking UvrB (lifetime of 304 ± 69 s; Nrepeats = 7 totaling nobs = 45,254 individual observations). f Mfd-YPet dissociates in 52 ± 4 s (Nrepeats = 8 totaling nobs = 36,486 individual observations) in the presence of UvrA(K37A) and UvrB, suggesting that ATP hydrolysis at the proximal site is required for promoting the dissociation of Mfd-YPet. Error bars are standard deviations from ten bootstrapped CRTDs. Source data are provided as a Source Data file.
