Fig. 4. Measurements of Mfd-YPet binding lifetimes in cells expressing mutant UvrB deficient in DNA loading or over-expressing wild-type UvrB.
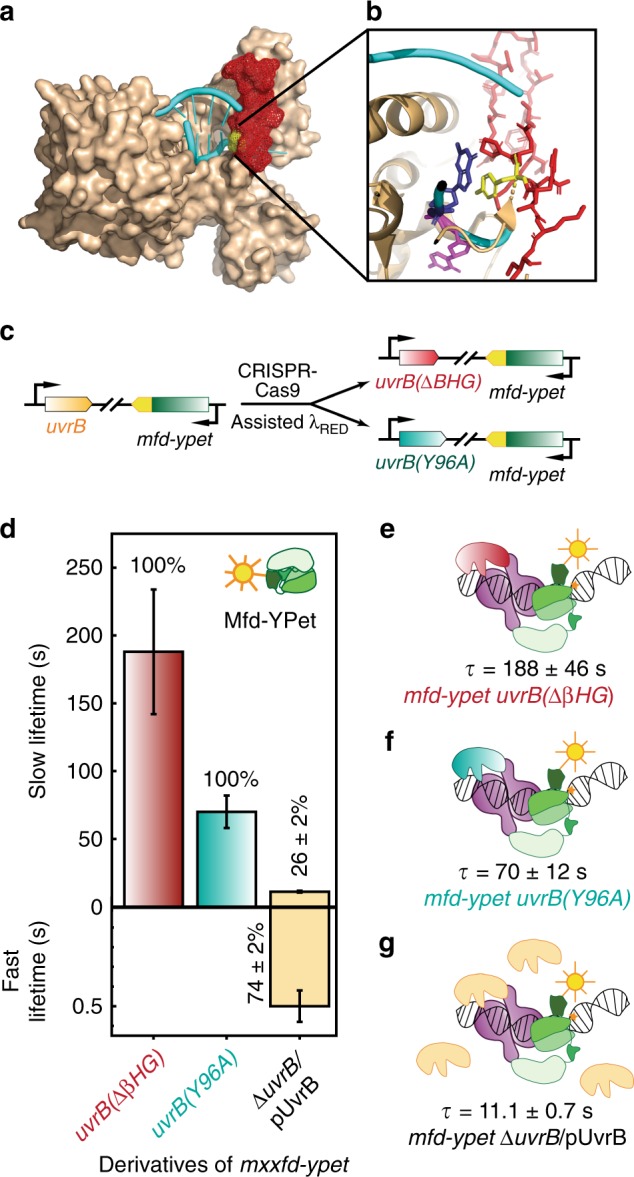
a Crystal structure of Bacillus caldotenax UvrB (brown) bound to a DNA hairpin (cyan) (PDB ID: 2FDC)39. The β-hairpin of UvrB is highlighted in red. b Zoomed-in view of the tyrosine residue Y96 (yellow) at the base of the β-hairpin (red). The tyrosine residue stabilises the DNA by forming π-stacking interactions with a guanine (blue) while a neighboring base (magenta) is flipped out into a hydrophobic pocket of UvrB. c Schematic of CRISPR-Cas9 assisted incorporation of deletions and point mutations in the uvrB gene. d Bar plots show lifetimes of DNA-bound Mfd-YPet in the corresponding genetic backgrounds. Where two kinetic sub-populations are detected, the fast lifetime is displayed in the lower panel. Numbers in percentages represent the amplitudes of kinetic sub-populations (see Supplementary Fig. 4). e Lifetime of Mfd-YPet in cells expressing mutant UvrB lacking the β-hairpin (red) was found to be 188 ± 46 s (Nrepeats = 9 totaling nobs = 37,950 individual observations), demonstrating this mutant UvrB fails to promote the dissociation of Mfd-YPet (green). f Similarly, mutant UvrB in which the absolutely conserved residue Y96 is replaced with alanine (cyan) also fails to promote the dissociation of Mfd-YPet (τMfd| uvrB(Y96A) = 70 ± 12 s; Nrepeats = 8 totaling nobs = 35,082 individual observations). g Overexpression of UvrB (orange) leads to a shorter lifetime of DNA-bound Mfd-YPet, compared to the constitutive level of UvrB (11.1 ± 0.7 s vs. 18 ± 1 s21). Nrepeats = 6 totaling nobs = 20,653 individual observations. Error bars are standard deviations from ten bootstrapped CRTDs. Source data are provided as a Source Data file.
