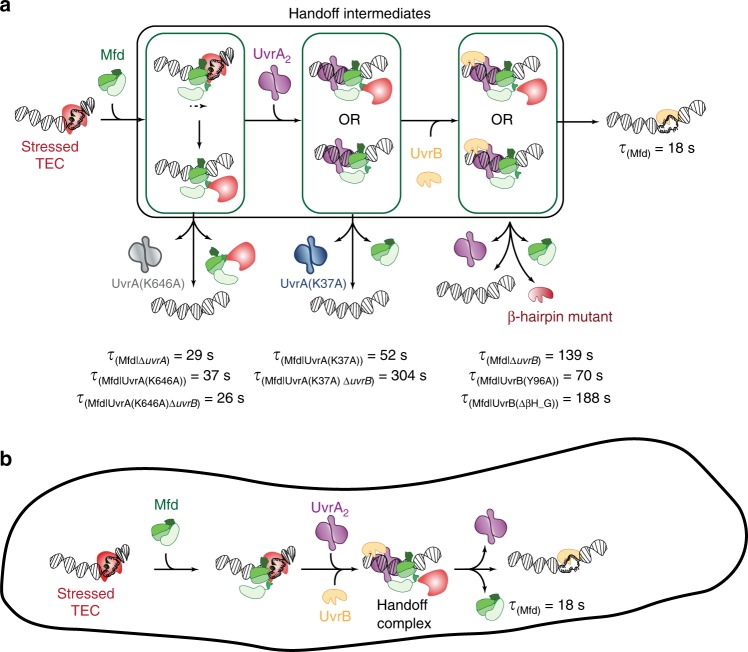
Fig. 5. Model for interactions of Mfd, UvrA and UvrB at sites of stressed TECs in cells.
a In cells, Mfd is normally auto-inhibited and does not stably interact with DNA. Binding to stressed TECs via its RNAP interacting domain leads to release of inhibition and a large conformational change in the protein. Following this, the motor domain engages DNA on the upstream edge of the transcription bubble and subsequent ATP hydrolysis leads to either transcription reactivation/termination. Mfd continues to translocate on dsDNA after remodeling the RNAP. Meanwhile, the UvrB homology module of Mfd is exposed permitting recruitment of UvrA. ATP hydrolysis at UvrA’s distal ATPase site is necessary for this engagement. Engagement with UvrA2 leads to arrest of translocation. Further, recruitment of UvrB (either sequentially as shown, or simultaneously with UvrA2) to the site leads to formation of the Mfd-UvrA-UvrA-UvrB handoff complex. Subsequent ATP hydrolysis at UvrA’s proximal ATPase site results in loading of UvrB on the DNA. Successful engagement of UvrB with the DNA via its β-hairpin is necessary for completing the handoff, culminating in dissociation of Mfd and UvrA2 from the site. b Facilitated dissociation model operates inside cells resulting in the simultaneous loss of UvrA2 and Mfd from the site of transcriptional stress.