Abstract
Candida albicans is a commensal fungus in humans, mostly found on the mucosal surfaces of the mouth, gut, vagina and skin. Incidence of ever increasing invasive candidiasis in immunocompromised patients, alarming occurrence of antifungal resistance and insufficient diagnostic methods demand more focused research into C. albicans pathogenicity. Consequently, in the present study, oleic acid from Murraya koenigii was shown to have the efficacy to inhibit biofilm formation and virulence of Candida spp. Results of in vitro virulence assays and gene expression analysis, impelled to study the protein targets which are involved in the molecular pathways of C. albicans pathogenicity. Proteomic studies of differentially expressed proteins reveals that oleic acid induces oxidative stress responses and mainly targets the proteins involved in glucose metabolism, ergosterol biosynthesis, lipase production, iron homeostasis and amino acid biosynthesis. The current study emphasizes anti-virulent potential of oleic acid which can be used as a therapeutic agent to treat Candida infections.
Subject terms: Mass spectrometry, Biofilms
Introduction
Candida is a genus of yeast with remarkable phenotypic characteristics and found as a commensal fungus in humans. Candida species are most commonly present in the genital tracts and other membrane tracts such as mucosal oral cavity, respiratory tract, gastrointestinal tract etc1. Although over hundred Candida species belong to this genus, C. albicans is responsible for the majority of Candida infection. Other medically important Candida species are Candida glabrata, Candida tropicalis, Candida dubliniensis, and Candida parapsilosis2. Also, C. albicans is able to form well structured, three dimensional biofilms comprising of round yeast cells, filamentous hyphae, pseudohyphae and exopolysaccharides which prevent the action of antifungal agents and safeguard the pathogen from host defense mechanism3. While most of the implant associated infections are caused by C. albicans biofilm, few non C. albicans Candida species (NCAC) including C. glabrata and C. tropicalis have also been reported for their participation in urinary tract and blood stream infections4. Worldwide, candidiasis is the fourth most healthcare associated infection in hospitalized patients and the pathogen Candida is well known for its device associated infection. In general, gold standard antifungal agents such as azoles, amphotericin B, polyenes and flucytosine are most frequently used for the treatment of Candida antifungal therapy. However, extensive usage of these antifungal agents makes the pathogen develop resistance against these drugs. In recent years, increased usage of these antimycotics in antifungal therapies, transplantation, AIDS and diabetes are the major factors of C. albicans infections in hospitalized patients. The pathogenic nature of Candida species is regulated by virulence traits such as morphological transition, contact sensing, biofilm development, invasion, adhesion on the cell surface and hydrolytic enzyme secretion5. To overcome these drug resistant and biofilm mediated Candida infections, there is an immediate requirement of alternative anti-pathogenic agents.
Though traditionally medicinal plants are extensively used for the treatment of many diseases, it is estimated that only 1–10% of ~250,000–500,000 plants on Earth are being used by humans6. In recent decades, medicinal plants have been widely reported for their antimicrobial effect against various bacterial and fungal pathogens. In addition, plants are also used as food preservatives, dietary supplements, food spoilage, flavor enhancers, etc. The major advantages of using plant-derived compounds as therapeutic agents are less adverse effects, multiple mode of action and low chances of antimicrobial resistance7. Our research group has recently reported the anti-infective potential of several phytocompounds against bacterial and fungal pathogens. For instance, 3-Furancarboxaldehyde and limonene against Group A Streptococcus8,9, 2-Furaldehyde diethyl acetal (Cocos nucifera), curcumin and other phytocompounds against Pseudomonas aeuginosa10,11, 3-O-methyl ellagic acid (Anethum graveolens) and vanillic acid (Actinidia deliciosa) against Serratia marcescens12,13, Embelin (Emblica ribes) in combination with ketoconazole against Malassezia spp.14, and synergistic combination of quinic acid and undecanoic acid against Candida spp15. In addition, oleic acid has been reported for its antibacterial and antifungal activity against various Gram positive and Gram negative bacterial pathogens and fungal pathogens16,17. However, reports are scanty on the mechanism of action of oleic acid. In this backdrop, the present study aimed to explore the anti-virulence efficacy of oleic acid derived from Murraya koenigii against Candida spp. through transcriptomic and proteomic approaches.
Results
Oleic acid disassembles Candida spp. biofilm
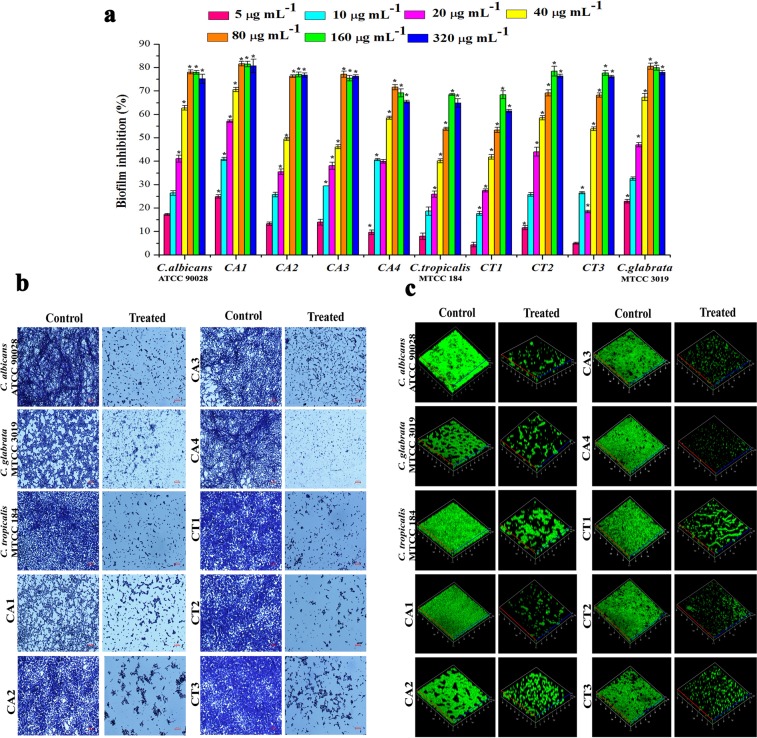
To investigate the effect of oleic acid on Candida spp. and to determine the biofilm inhibitory concentration (BIC), standard crystal violet quantification method was used. The results of antibiofilm assay showed a concentration dependent increase in biofilm inhibition. BIC of oleic acid was varying between Candida species. For the wild type (ATCC 90028) and clinical isolates of C. albicans (CA1, CA2, CA3 and CA4) and C. glabrata (MTCC 3019), BIC was found to be 80 µg mL−1 (Fig. 1a). Whereas, BIC of C. tropicalis (MTCC184), and the clinical isolates (CT1, CT2 and CT3) was found to be 160 µg mL−1.
Figure 1.
Antibiofilm activity of oleic acid against Candida spp. without affecting fungal growth and viability. (a) Biofilm inhibitory potential of oleic acid against Candida spp. in a dose dependent manner in spider broth at 37 °C for 24 h. (b) Light microscopic images depicting Candida spp. biofilm formed on glass surfaces in the absence and presence of oleic acid in spider broth at 37 °C for 24 h. Magnification 400×, scale bar – 200 µm. (c) 3D representation of biofilm dispersal induced by oleic acid in Candida spp. as visualized by confocal laser scanning microscopy.
Oleic acid disassembles Candida spp. biofilm without affecting fungal growth and viability
Oleic acid did not inhibit the growth of all the tested Candida strains at varying concentrations (10 µg mL−1 to 640 µg mL−1) confirming the non-fungicidal antibiofilm effect of oleic acid (Supplementary Fig. S1a). Also, this result was further authenticated by XTT assay wherein the viability of oleic acid treated cells was similar to that of control samples in all the tested strains (Supplementary Fig. S1b).
Microscopic visualization confirms biofilm disassembly of oleic acid
To evaluate the antibiofilm efficacy of oleic acid on glass surface, light microscopic analysis was performed. In the light micrographs, control samples were seen with thick layer of biofilm matrix consisting of yeast and hyphae cells. Whereas microcolony formation and hyphal elongation was completely inhibited by oleic acid in treated samples (Fig. 1b). To further corroborate the antibiofilm activity of oleic acid, CLSM analysis was performed. Three-dimensional view of Candida spp. biofilm clearly revealed reduction in the biofilm thickness and biomass and mature biofilm architecture in all the tested slides when compared to their respective controls (Fig. 1c).
Oleic acid controls filamentous growth of Candida spp
Effect of oleic acid on Candida spp. filamentation was assessed using spider agar supplemented with 10% FBS. In the case of C. albicans wild type strain (ATCC 90028) and the clinical isolates (CA1, CA2, CA3 and CA4), filamentous growth was significantly inhibited upon oleic acid treatment (Fig. 2a). On the other hand, in C. tropicalis (MTCC) and its isolates (CT1, CT2 and CT3) only slight inhibition in the hyphal growth was noticed. C. glabrata is hyphal negative, thus no filamentous growth was observed even after 72 h of incubation.
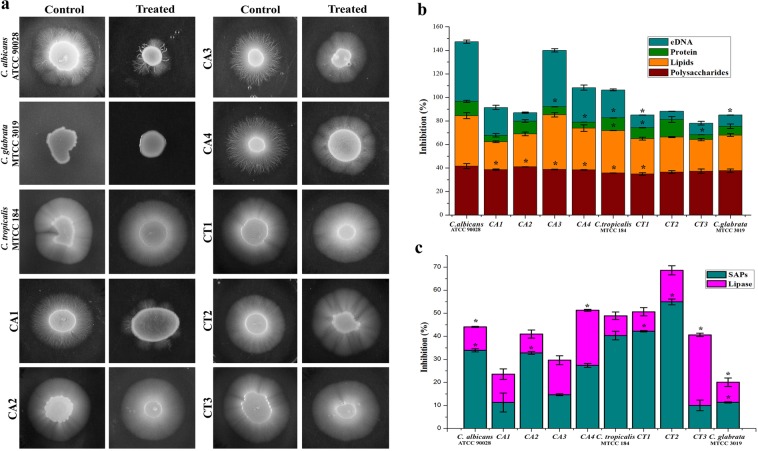
Figure 2.
(a) Oleic acid inhibiting filamentous growth of Candida spp. in spider agar supplemented with 10% FBS. Oleic acid strongly reduced the filamentation ability in C. albicans (ATCC 90028) and clinical isolates (CA1, CA2, CA3 and CA4). Moderate reduction in the filamentous growth was observed in C. tropicalis (MTCC 184) and its clinical isolates (CT1, CT2, CT3). No filamentous growth was observed in C. glabrata (MTCC 3019). (b) Quantification of EPS components such as polysaccharides, lipids, proteins and eDNA extracted from Candida spp. biofilm matrix by phenol sulfuric acid method (absorbance at 490 nm), phospho-vanillin method (absorbance at 545 nm), Bradford method (absorbance at 595 nm) and nano spectrophotometer (absorbance at 260/280 ratio), respectively. Oleic acid mainly inhibits the polysaccharides and lipids present in the biofilm matrix. (c) Effect of oleic acid on secreted aspartyl proteinases (SAPs) and lipases produced by Candida spp. using bovine serum albumin agar and lipase agar, respectively. Error bars represent standard deviations from the mean (n = 3). Statistical significance was analyzed using one way ANOVA-Duncan’s post-hoc test and single asterisk represent p < 0.05.
Oleic acid disperses biofilm matrix enclosed by extra polymeric substances (EPS)
The EPS of Candida spp. encompasses three-dimensional structure of yeast, hyphae, pseudohyphae and biomolecules such as carbohydrates, proteins, lipids and nucleic acids15. In all the tested Candida strains, polysaccharides and lipids present in the EPS were decreased up to 35–41% and 26–47%, respectively (Fig. 2b). Moreover, slight inhibition (4–14%) was observed in the protein level of EPS upon oleic acid treatment. Intriguingly, oleic acid was found to inhibit eDNA level of EPS in an uneven manner, wherein a maximum of 47% inhibition was observed in CA3 and only 6% inhibition was noticed in CT3 strain.
Oleic acid reduces secreted hydrolases production in Candida spp
Effect of oleic acid on Candida spp. secreted aspartyl proteinases (SAPs) and lipase production was qualitatively measured using BSA and tributyrin agar, respectively15. White precipitation zone around the colonies indicates the SAPs production. Zone diameter was measured for both control and treated plates and the percentage of inhibition was calculated. Likewise, lipase production was also quantitatively measured by zone of clearance and the percentage of inhibition was measured. In all the tested strains, production of SAPs and lipases was inhibited up to 10–54% and 8–30%, respectively (Fig. 2c).
Oleic acid modifies ergosterol constituent of fungal cell membrane
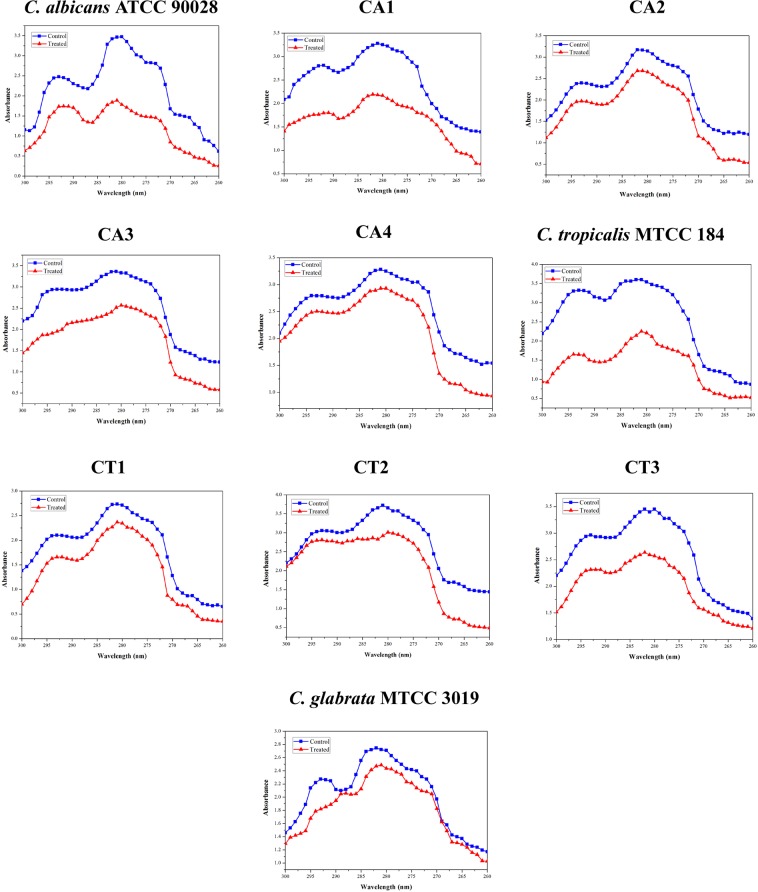
Ergosterol is an important component of fungal cell membrane maintaining the integrity, permeability and structure3. Hence, the change in the ergosterol production upon treatment with oleic acid was evaluated using UV spectrophotometer. UV scanning spectra represented four peaks between 260–300 nm corresponding to ergosterol and sterol intermediates and the flat line without peaks indicated the absence of sterol. As can be seen from Fig. 3, Oleic acid drastically reduced the ergosterol content of C. albicans, C. tropicalis, CA1, CA3 and CT3 strains and in the remaining Candida strains moderate ergosterol inhibition was noticed.
Figure 3.
UV spectra revealing the effect of oleic acid on ergosterol profile of Candida spp. Ergosterol present in Candida spp. with and without oleic acid scanned from 260 to 300 nm. Reduction in the peak height represents changes in ergosterol content. UV scanning spectra represented four peaks between 260–300 nm corresponding to ergosterol and sterol intermediates.
Effect of oleic acid on C. albicans adhesion ability and H2O2 sensitivity
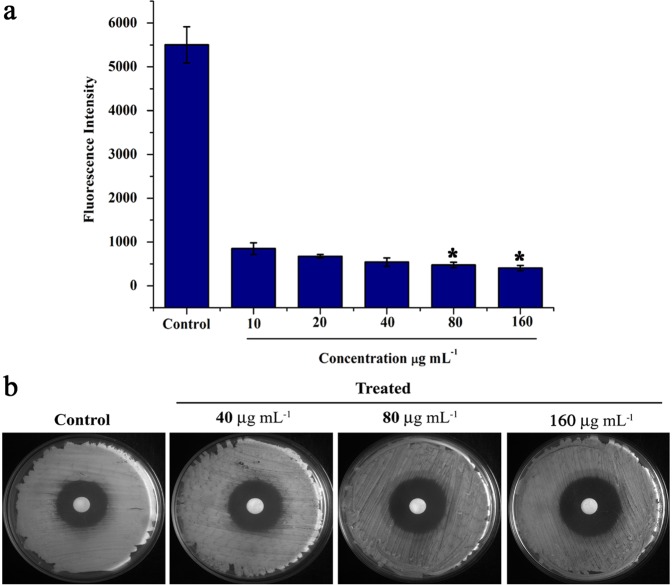
The very first event of Candida biofilm formation is the adhesion to host surfaces for both colonization and establishment of infections. Thus, effect of oleic acid on C. albicans adherence to the polystyrene surfaces was assessed using alamar blue assay. The fluorescent intensity of alamar blue or resazurin dye was found to be decreased in a dose dependent manner in oleic acid treated samples when compared to control. This result clearly depicts the anti-adhesion ability of oleic acid on C. albicans biofilm attached to the polystyrene surfaces (Fig. 4a).
Figure 4.
Effect of oleic acid C. albicans adhesion ability and H2O2 sensitivity. (a) Alamar blue reduction assay was performed to check the effect of oleic acid on C. albicans adhesion ability to the polystyrene surfaces. Fluorescence intensity dose dependently decreased in oleic acid treated samples compared to control. (b) H2O2 sensitivity assay - Representative YEPD agar plates depicting the effect of oleic acid treatment on the sensitivity of C. albicans to H2O2. Error bars represent standard deviations from the mean (n = 3). Statistical significance was analyzed using one way ANOVA-Duncan’s post-hoc test and single asterisk represent p < 0.05.
Results of H2O2 sensitivity assay showed that oleic acid treated C. albicans cells were more sensitive to H2O2 than control. Briefly, in control plates, zone of clearance around H2O2 was measured as 32 mm (Fig. 4b), whereas in the case of 40, 80 and 160 µg mL−1 of oleic acid treated plates, sensitivity to H2O2 were increased with the diameter of 34, 36 and 38 mm respectively (Fig. 4b).
Impact of oleic acid on C. albicans virulence gene expression
Quantitative PCR was used to assess the inhibitory effect of oleic acid on the transcriptional regulatory network of biofilm formation and virulence of C. albicans. Among the tested genes, candidate genes involved in adhesion (als1), SAPs production (sap2), hyphal elongation (hwp1) and filamentation (cst20) were significantly down regulated up to 8.9, 4.1, 5.6 and 4.4 fold respectively. In addition, genes involved in ergosterol production (erg11 – 2.5 fold), cell adhesion (ras1 – 3.3 fold) and hyphal transcription factor (cph1- 1.3 fold) were moderately down regulated by oleic acid. Besides, oleic acid slightly down regulated (<1.0 fold) the expression of other genes. But, the regulation of cell wall related genes chitin synthase-3 (chs3 – 1.0 fold) and chitinase (cht4 – 0.9 fold) were found to be slightly upregulated (Fig. 5).
Figure 5.
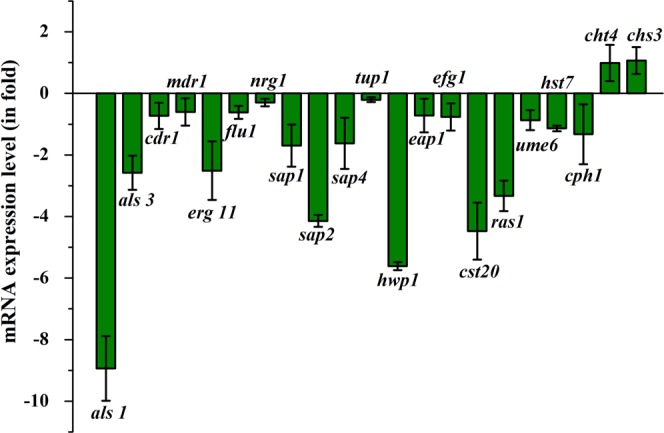
Gene expression profile of C. albicans. Impact of oleic acid on the expression of candidate genes involved in virulence factors (als1, als3 - adhesion, cdr1, mdr1 – efflux pump mechanism, erg11 - ergosterol production, flu1 – fluconazole resistance gene, sap1, sap2, sap4 - SAPs production, hwp1 - hyphal elongation, nrg1, tup1 – negative regulators of transcription, cst20, hst7 – filamentation, efg1, cph1 – transcription factor, ume6 - Transcriptional regulator of filamentous growth, ras1, eap1 – cell adhesion, chs3 – synthesis of chitin, cht4 – degradation of chitin). Relative gene expression level was determined using the ΔΔCT method. Error bars represent standard deviations from the mean (n = 3).
Effect of oleic acid treatment on the cellular proteome of C. albicans
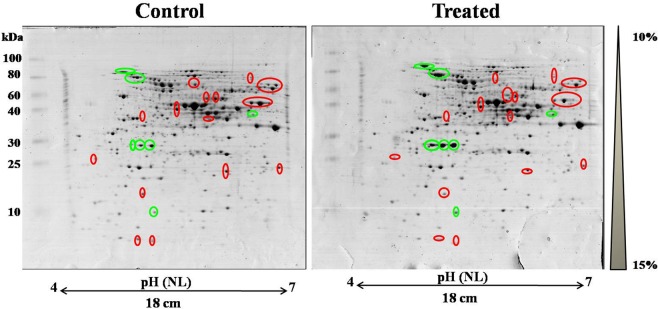
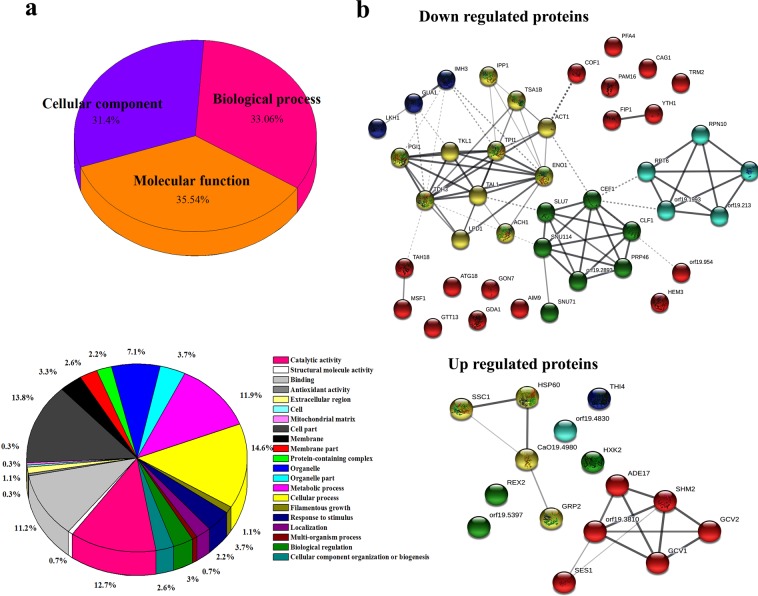
To find out the mechanism of action of oleic acid on C. albicans virulence, two dimensional gel electrophoresis (2D-PAGE) coupled with mass spectrometry (MALDI-TOF/TOF) technique was used. Intracellular proteins from C. albicans grown (16 h) in the absence and presence of oleic acid were extracted by sonication. The extracted proteins were quantified using standard Bradford method and SDS-PAGE was performed in order to check the quality of protein and identify the differential expression between control and treated samples. Afterwards, cellular proteome of control and oleic acid treated C. albicans was assessed using 2D-PAGE (Fig. 6). Image Master Platinum software (Version- 7.0, GE Healthcare, USA) was used to detect and match the protein spots present in the control and treated gels. Based on the results obtained from densitometric analysis, among the 300 detected spots, eleven and forty spots were found to be up regulated and down regulated (>1.5 fold), respectively. Based on statistical significance (ANOVA - P < 0.05) and differential expression (≥1.5 fold), 51 spots were selected for protein identification and subjected to MALDI-TOF/TOF analysis. Then, the obtained MALDI spectra were matched with Swissprot.2017.11.01 database using MS-Fit online software tool. The details of differentially regulated proteins and their function, number of peptides matched, sequence coverage and MOWSE score are listed in Table 1. In addition, gene ontology analysis in UniProt was used to identify the functions of differentially regulated proteins. From this analysis it became evident that the differentially expressed proteins are mainly involved in cellular process (37%), catalytic activity (33%), metabolic process (30%), binding (29%), response to stimulus (9%), biological regulation (8%), and cellular component organization (7%) (Fig. 7a). Furthermore, protein-protein interaction among differentially regulated proteins (both up regulated and down regulated proteins) was done using STRING v.11 database with confidence score (0.4) and the resultant protein-protein interaction map is shown in Fig. 7b. These results suggest that the differentially regulated proteins are predominantly involved in major pathways including proteasome, spliceosome, carbon metabolism, biosynthesis of antibiotics, biosynthesis of secondary metabolites and metabolic pathways.
Figure 6.
Effect of oleic acid on intracellular proteome of C. albicans. Representative gel images depicting the comparison between intracellular protein extract of C. albicans grown in the absence and presence of 80 µg mL−1 of oleic acid by 2- D gel electrophoresis. One thousand microgram of protein was subjected to isoelectric focusing and resolved in 10–15% gradient SDS-PAGE and the protein spots were stained with MS compatible CBB G-250. Up regulated and down regulated protein spots are highlighted in green and red circles, respectively.
Table 1.
List of differentially expressed proteins of C. albicans treated with oleic acid identified using MALDI-TOF/TOF.
| Spot ID | Fold | P-value | Accession No. | Description | MOWSE score | Coverage (%) | No. of Peptides matched | Gene Name |
|---|---|---|---|---|---|---|---|---|
| Down regulated proteins | ||||||||
| CAP1 | 4.2 | 0.002 | P83773 | Acetyl-CoA hydrolase | 2.86E + 11 | 19.7 | 20 | ACH1 |
| CAP3 | 3.2 | 0.006 | P43067 | Alcohol dehydrogenase | 184 | 6.3 | 7 | ADH1 |
| CAP5 | 2.8 | 0.003 | O94048 | Porphobilinogen deaminase | 1195 | 16.8 | 12 | HEM3 |
| CAP6 | 2.8 | 0.152 | Q59T36 | mRNA 3′-end-processing protein | 6.41E + 06 | 30 | 15 | YTH1 |
| CAP8 | 2.6 | 0.003 | O74713 | High-affinity glucose transporter | 4413 | 8.6 | 10 | HGT1 |
| CAP10 | 2.4 | 0.036 | Q59W04 | EKC/KEOPS complex subunit | 10310 | 33.1 | 7 | GON7 |
| CAP11 | 2.4 | 0.001 | Q5APF2 | GMP synthase [glutamine-hydrolyzing] | 32644 | 20.6 | 9 | GUA1 |
| CAP12 | 2.3 | 0.004 | P34725 | Phospho-2-dehydro-3-deoxyheptonate aldolase, phenylalanine-inhibited | 1382 | 8.2 | 6 | ARO3 |
| CAP16 | 2.0 | 0.006 | Q5A4Z2 | Uncharacterized protein | 1025 | 21 | 13 | orf19.954 |
| CAP17 | 2.0 | 0.238 | Q92211 | Glyceraldehyde-3-phosphate dehydrogenase | 39759 | 23 | 12 | TDH1 |
| CAP19 | 2.0 | 0.002 | Q0ZIB4 | Mitochondrial GTPase elongation factor Tu (Fragment) | 902 | 32.6 | 4 | TUF1 |
| CAP20 | 2.0 | 0.000 | Q5AM84 | U1 small nuclear ribonucleoprotein component | 952 | 8.1 | 12 | SNU71 |
| CAP21 | 2.0 | 0.044 | Q8TGH6 | Guanosine-diphosphatase | 489 | 13.4 | 8 | GDA1 |
| CAP24 | 1.9 | 0.000 | A0A1D8PMW6 | Cofilin | 3093 | 59.6 | 8 | COF1 |
| CAP26 | 1.9 | 0.000 | P28868 | Guanine nucleotide-binding protein | 1943 | 9.3 | 7 | CAG1 |
| CAP29 | 1.8 | 0.043 | Q5AD28 | Phenylalanine–tRNA ligase | 94591 | 17.7 | 9 | MSF1 |
| CAP30 | 1.8 | 0.000 | Q59X29 | Proteasome regulatory particle base subunit | 1204 | 11.1 | 4 | RPN10 |
| CAP31 | 1.8 | 0.130 | Q02751 | Alpha-glucosidase | 4802 | 19.8 | 7 | MAL2 |
| CAP32 | 1.8 | 0.000 | A0A1D8PCG3 | Trm2p | 46991 | 16 | 10 | TRM2 |
| CAP34 | 1.7 | 0.004 | Q0P7I3 | Dit1 protein | 661 | 11.2 | 7 | DIT1 |
| CAP36 | 1.7 | 0.052 | Q5ADL4 | Pre-mRNA-splicing factor | 1.92E + 06 | 15.7 | 12 | SLU7 |
| CAP37 | 1.7 | 0.002 | Q5A922 | Altered inheritance of mitochondria protein 9 | 2030 | 14.9 | 10 | AIM9 |
| CAP38 | 1.7 | 0.000 | Q8J224 | Alpha subunit of farnesyl transferase | 210 | 14.7 | 4 | RAM2 |
| CAP40 | 1.7 | 0.001 | P83777 | Inorganic pyrophosphatase | 984 | 14.6 | 6 | IPP1 |
| CAP44 | 1.6 | 0.002 | Q5A750 | Transketolase | 836518 | 15.7 | 10 | TKL1 |
| CAP46 | 1.6 | 0.007 | Q5AD27 | NADPH-dependent diflavin oxidoreductase 1 | 14468 | 13.1 | 8 | TAH18 |
| CAP47 | 1.6 | 0.001 | G1UA34 | Putative uracil phosphoribosyltransferase | 5054 | 33.5 | 10 | FUR1 |
| CAP48 | 1.6 | 0.002 | P78594 | Cytosine deaminase | 90713 | 11.3 | 6 | FCA1 |
| CAP50 | 1.6 | 0.015 | A0A1D8PJQ3 | Gtt13p | 10203 | 12.2 | 11 | GTT13 |
| CAP51 | 1.6 | 0.018 | Q5ABA6 | Autophagy-related protein 18 | 12699 | 17.2 | 10 | ATG18 |
| CAP53 | 1.6 | 0.006 | Q59ZW9 | Mitochondrial import inner membrane translocase subunit | 2059 | 22.3 | 6 | PAM16 |
| CAP54 | 1.6 | 0.004 | A0A1D8PFR4 | Actin | 91474 | 26.1 | 12 | ACT1 |
| CAP55 | 1.6 | 0.048 | G1UAT0 | Uncharacterized protein | 1676 | 9.1 | 12 | CaJ7.0419 |
| CAP58 | 1.5 | 0.001 | P78589 | Squalene synthase | 502 | 12.3 | 10 | ERG9 |
| CAP59 | 1.5 | 0.002 | Q59RQ6 | Dihydrolipoyl dehydrogenase | 203000000 | 21.2 | 16 | LPD1 |
| CAP60 | 1.5 | 0.000 | Q59NB8 | Leukotriene A-4 hydrolase homolog | 576307 | 15.4 | 13 | LKH1 |
| CAP62 | 1.5 | 0.002 | Q5AGV7 | Palmitoyltransferase | 56899 | 11.9 | 11 | PFA4 |
| CAP63 | 1.5 | 0.046 | O94091 | Lipase 1 | 1808 | 10.9 | 5 | LIP1 |
| CAP64 | 1.5 | 0.067 | Q9HFV5 | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase | 5199 | 15.4 | 6 | HIS6 |
| CAP65 | 1.5 | 0.003 | P0CU34 | Peroxiredoxin | 36223 | 25.5 | 7 | TSA1B |
| Up regulated proteins | ||||||||
| CAP2 | 3.2 | 0.001 | Q5A313 | Uncharacterized protein | 580 | 9.7 | 7 | CAALFM_CR08740WA |
| CAP4 | 3.0 | 0.000 | A0A1D8PGG3 | Rex2p | 23793 | 27.8 | 12 | REX2 |
| CAP7 | 1.9 | 0.410 | O13426 | Serine hydroxymethyltransferase | 7.85E + 03 | 18.7 | 8 | SHM2 |
| CAP13 | 2.3 | 0.002 | C4YNP4 | Thiamine thiazole synthase | 2.50E + 10 | 37 | 22 | THI4 |
| CAP15 | 2.1 | 0.002 | Q5A7S0 | Golgi transport complex subunit | 4491 | 16.9 | 9 | orf19.5397 |
| CAP25 | 1.9 | 0.017 | P83776 | Hexokinase-2 | 732657 | 27.7 | 13 | HXK2 |
| CAP28 | 1.8 | 0.002 | Q9HGT6 | Serine–tRNA ligase, cytoplasmic | 375 | 13.2 | 11 | SES1 |
| CAP43 | 1.6 | 0.052 | Q5APD5 | Uncharacterized protein | 28945 | 18.2 | 5 | orf19.4830 |
| CAP45 | 1.6 | 0.000 | G1UAH4 | Uncharacterized protein | 1405 | 20.8 | 14 | CaJ7.0423 |
| CAP52 | 1.6 | 0.037 | P41797 | Heat shock protein SSA1 | 5056 | 26.5 | 13 | SSA1 |
| CAP67 | 1.5 | 0.022 | P83784 | Heat shock protein SSC1 | 390937 | 17.3 | 11 | SSC1 |
Figure 7.
Gene ontology and STRING analysis of differentially regulated proteins of C. albicans upon oleic acid treatment. (a) Gene ontology analysis of differentially regulated (both up regulated and down regulated) proteins using UniProt database. The differentially expressed proteins are mainly involved in cellular process (37%), catalytic activity (33%), metabolic process (30%), binding (29%), response to stimulus (9%), biological regulation (8%), and cellular component organization (7%) (b) Protein-protein interaction prediction map of (i) down regulated and (ii) up regulated proteins of C. albicans obtained using STRING v.11 database (confidence score 0.400). The interaction map shows the proteins involved in interrelated pathways such as proteasome, spliceosome, carbon metabolism, biosynthesis of antibiotics, biosynthesis of secondary metabolites and metabolic pathways. Filled protein nodes that signify the availability of protein 3D structural information is known or predicted.
Discussion
Historically, Candida spp., Cryptococcus spp., Aspergillus spp. and Fusarium spp. have been reported as opportunistic fungal pathogens in hospitalized patients18. Candidiasis is the recurrent fungal infection caused by Candida genus, primarily C. albicans followed by C. parapsilosis, C. glabrata, C. tropicalis and C. krusei19. Although, emergence of antifungal resistance to conventional antifungal drugs is the major challenge to research scientists and pharmaceutical companies20. Recently published reports have suggested that plant derived compounds or phytocompounds can certainly be more efficient in controlling biofilm related infections21–23. Furthermore, the chances of drug resistance and adverse effects of these plant derived compounds are very low than conventional antibiotics or antifungal agents12. In this context, the present study established the inhibitory potential of oleic acid derived from M. koenigii against Candida spp. biofilm and virulence through in vitro assays, gene expression studies and proteomic approaches.
Primarily, Biofilm inhibitory concentration (BIC) of oleic acid was determined for all the tested Candida strains and exhibited maximum of 80% biofilm inhibition. Likewise, oleic acid derived from Withania somnifera and oleic acid coated magnetic nanoparticles exhibited substantial antibiofilm activity against various bacterial pathogens24,25. Previous studies revealed the antimicrobial activity of oleic acid against certain bacterial and fungal pathogens17,26. However, according to Rasmussen and Givskov (2006)27–29, compounds or antibiotics that kill or inhibit the growth of microorganism situates an organism under selection pressure to develop drug resistance. Hence, antifungal effect of oleic acid was assessed using broth dilution assay and the results clearly suggested that oleic acid did not inhibit the growth of Candida spp. XTT assay was performed to corroborate the non-fungicidal effect of oleic acid against Candida spp. The obtained results manifestly proved that the oleic acid does not inhibit the metabolic viability of Candida strains. Besides, microscopic techniques such as light microscopy and CLSM further demonstrated the inhibitory effect of oleic acid on Candida spp. biofilm formation and yeast to hyphal transition. It is akin to mammalian apoptosis inducer BH3l-1 and its derivatives effectively blocking the yeast-to-hyphal and yeast-to-pseudohyphal transitions in C. albicans30.
In Candida spp., filamentous growth is influenced by several factors including glucose and nitrogen starvation, oxygen deficiency, presence of N-acetylglucosamine and serum31. The filamentation assay results evidently showed that oleic acid significantly inhibits the filamentous growth of wild type C. albicans and C. tropicalis. However, trivial inhibition was observed in the filamentous growth of clinical isolates of Candida spp. This result is in agreement with an earlier study made by Manoharan et al.32, wherein, 7-benzyloxyindole (indole derivative) considerably inhibited the filamentous growth of C. albicans even after 10 days of incubation. In the case of Candida biofilm, EPS matrix furnishes protective environment to the biofilm cells from host immune system and from the antifungal agents3,15. Hence, quantification of EPS components was carried out which revealed that oleic acid treatment could significantly reduce the carbohydrates, lipids and eDNA content of EPS. However, no considerable change was detected in the protein level of EPS. In the same way, compounds from natural resources such as 2,4-di-tert-butylphenol from Vibrio alginolyticus G16, usnic acid from lichen and 5-hydroxymethyl-2-furaldehyde from Bacillus subtilis efficiently disrupted the EPS of C. albicans33–35. Secreted hydrolases such as SAPs, phospholipases and lipases play a significant role in Candida spp. pathogenicity and these enzymes facilitate the invasion of hyphal cells into host tissues36,37. SAPs and lipase production of Candida spp. were qualitatively measured, while oleic acid moderately inhibited SAPs production. Previously, mycogenic AgNPs have been shown to exhibit significant inhibition against SAPs production and biofilm growth in C. albicans and NCAC species38. On the other hand, only slight inhibition was noticed in lipase production. Enzymes involved in ergosterol pathway are the main target of most of the antifungal agents including azoles and polyenes. Besides ergosterol, membrane lipid modulates cell membrane fluidity, permeability, and integrity15,39,40. In the tested Candida strains, oleic acid produced substantial changes in the ergosterol content of each Candida spp. which undoubtedly suggest that oleic acid could be a potent drug than conventional antifungal agents. Similarly, Masood et al.41 have reported that a novel series of 1,2,3-triazole–quinazolinone conjugates interferes with sterol biosynthetic pathway and decreases the ergosterol content in C. albicans. Adhesion to biotic or abiotic surfaces is the first step of C. albicans biofilm formation15. Results of alamar blue assay clearly indicate that oleic acid significantly inhibited the adhesion ability of C. albicans cells to the polystyrene surfaces in a concentration dependent manner. In vitro assay results further substantiated at the transcription level using real time PCR. Oleic acid treatment considerably down regulated the genes involved in virulence of Candida spp., such as adhesion (als1, als3, eap1), ergosterol biosynthesis (erg11), SAPs production (sap1, sap2, sap4), filamentation (hwp1, efg1, cst20, ras1, ume6, hst7) and efflux pump mechanism (cdr1, mdr1). In a similar way, synergistic combination of quinic acid and undecanoic acid significantly down regulated the major virulence genes of C. albicans15. Yet another study reported that linalool, terpene alcohol, could inhibit yeast-hyphal transition through cAMP-PKA pathway (via efg1 gene), MAPK pathway (via cph1 gene), control adhesins (als3 gene) and hyphal maintanence (eed1, ume6, hgc1) thereby persuading the C. albicans biofilm formation42. Conversely, oleic acid did not show any negative impact on the genes chitin synthase-3 (chs3) and chitinase-4 (cht4) which are essential for cell wall strength and flexibility, respectively43.
In the current study, 51 proteins were identified by MALDI-TOF/TOF followed by MS-Fit analysis. Based on proteins’ function and their importance in C. albicans virulence, 20 differentially regulated proteins were discussed in this study. Proteomic analysis revealed that oleic acid significantly down regulated the expression of acetyl-coA hydrolase (Ach1p) by 4.2 fold. pH homeostasis plays an important role in virulence mechanism of C. albicans in various host environments and the pathogen is capable of changing acidic/alkaline environment to neutral pH. Neutral pH promotes the morphological transition of yeast to hyphae which enhance the virulence of pathogen. This sort of pH adaptation is related to aminoacids catabolism by the enzyme acetyl-coA hydrolase and urea amidolyase (Dur1)44,45. The down regulation of Ach1p by oleic acid is comparable to physiological assays, wherein, microscopic visualization and filamentation assay results clearly proved that oleic acid considerably inhibited the hyphal growth of C. albicans in spider medium (at pH 7.2).
Alcohol dehydrogenase enzyme (Adh1) catalyzes both ethanol fermentation and metabolism. Adh1 is one of the factors which controls planktonic growth and biofilm formation of C. albicans through an ethanol dependent mechanism. Further, Adh1 also regulates other virulence factors including yeast-to-hyphal transition, drug efflux mechanism46–50. In the present study, Adh1p expression was down regulated up to 3.2fold upon treatment with oleic acid. In C. albicans, there are three known iron uptake systems including iron utilization from hemoglobin27,51,52. Further, porpholinogen deaminase (Hem3) is an enzyme involved in heme biosynthetic pathway which gets induced in the presence of high iron and CO2 and the same was down regulated upto 2.8 fold by oleic acid treatment. Likewise, porpholinogen deaminase protects Aspergillus nidulans from reactive nitrogen species or nitrosative stress53.
In C. albicans, glucose majorly influences the yeast-to-hyphal transition and other virulence factors such as adhesion, biofilm formation, oxidative stress resistance, invasion, and antifungal drug tolerance54,55. Moreover, Glucose transporters are considered as major targets for antimicrobial drug development. In the present study, oleic acid treatment down regulated the expression of Hgt1 protein (2.6 fold) in YEPD + FBS medium. GMP synthase [glutamine-hydrolyzing] enzyme (GUA1) is involved in the subpathway of purine metabolism which synthesizes guanine 5’-monophosphate (GMP) from xanthine 5’-monophosphate (GMP). Earlier reports suggested that Gua1 could be a potent target for an antifungal compound which is essential for growth and virulence of C. albicans. An in vivo study results in murine model of systemic candidiasis clearly suggested that heterozygous mutant of Gua1 strain (gua1/gua1) was completely avirulent56–58. Down regulation of Gua1p in the current study clearly confirmed that oleic acid modulates the pathogenicity of C. albicans.
Phospho-2-dehydro-3-deoxyheptonate aldolase, phenylalanine-inhibited (Aro3), an enzyme involved in aromatic amino acid biosynthesis, is down regulated (2.3 fold) by oleic acid. Yin et al.59, reported the comparative response of general amino acid responses (GCN) of S. cerevisiae and C. albicans at the proteomic level. From the results, it is clear that Aro3p is increased in response to the histidine analogue, 3-aminotriazole, though oleic acid down regulated the expression of Aro3p. This report clearly suggests that C. albicans cells use amino acids as nitrogen source for their growth in host cells and amino acids utilization directly influences fungal pathogenesis by activating virulence factors such as biofilm growth and hyphal morphogenesis60,61. In the present study, oleic acid inhibited the hyphal morphology and biofilm formation, accordingly down regulated the expression of Aro3p.
In glycolysis pathway, glyceraldehyde-3-phosphate dehydrogenase or GAPDH (Tdh1) converts D-glyceraldehyde 3-phosphate into pyruvate. In previous report, Tdh1p expression was found to be up regulated in the presence of N-acetylglucosamine (GlcNAc) which induces morphological transition at amino acid depleted condition62. This enzyme Tdh1p expression was found to be 2.0 fold down regulated upon treatment with oleic acid. Guanosine diphosphatase or GDPase encoded by GDA is an enzyme involved in nucleotide sugar transport/antiport cycle from golgi apparatus to cytosol and it is present in yeast and hyphal forms of C. albicans. Null mutant strain gda1/gda1 of C. albicans is severely defective in O-mannosylation and cell wall phosphate content which can slightly affect the hyphal growth63–65. Hyphal inhibitory effect of oleic acid could be the reason for the down regulation of Tdh1p and Gda1p.
In the present study, oleic acid down regulated the expression of alpha glucosidase (Mal2p) by 1.8 fold in glucose containing medium. Alpha glucosidase is an enzyme which hydrolyzes sucrose for sucrose utilization and is induced by maltose and suppressed by glucose. In 2006, Bramono et al.66 reported that cell surface alpha-glucosidase activity was very high in C. tropicalis, C. albicans and C. parapsilosis than other Candida species. Recently, Kim and his colleagues (2018)67 reported that Magnoflorine compound could inhibit the alpha-glucosidase activity (>50%) in C. albicans which is essential for normal cell wall composition and virulence.
Thiamine dependent enzyme, transketolase (Tkl1p), an important enzyme of carbon metabolism, is up regulated in response to cadmium, salt stress and hydrogen peroxide68. In recent years targeting thiamine biosynthetic pathways has emerged as the new strategy for the development of antifungal drugs (Meir and Osherov, 2018)69. Also, Siemieniuk, et al.70 reported the inhibitory effect of thiamine antivitamins on the growth and metabolism of pathogenic C. albicans and M. pachydermatis. In the present study, down regulation in the expression of Thiamine dependent enzyme, transketolase (Tkl1p), suggests that oleic acid may possibly target the thiamine biosynthetic pathway for its antivirulence property.
NADPH-dependent diflavin oxidoreductase 1 (Tah18p) is the part of iron-sulfur (Fe-S) cluster assembly, required for the maturation of extra mitochondrial Fe-S protein. In C. albicans, iron is an important micronutrient which controls its pathogenicity where survival of this pathogen in host depends on iron homeostasis by regulating iron uptake and storage71. In addition, C. albicans consist of tripartite system of transcription factors such as Sef1 (Zn2Cys6 DNA-binding protein), Sfu1 (GATA factor), and Hap43 (CCAAT binding complex) to control iron homeostasis72. These transcriptional regulators (Sef1 and Rim101) activate the virulence genes expression during iron deprivation. In the present study, oleic acid down regulated the Tah18p, thereby inactivating the Fe-S assembly and downregulated the expression of virulence factors.
Mitochondrial import inner membrane translocase subunit Tim16 (Pam16) is a part of PAM complex which is involved in the ATP-dependent translocation of the transit peptide containing proteins. In addition to that, PAM16 gene is involved in carbohydrate metabolism and up regulated during intermediate and mature biofilm development stages73. In the current study, oleic acid inhibited mature biofilm formation without affecting metabolic viability, which is also reflected on the down regulation of Pam16p (1.6 fold).
Squalene synthase (Erg9) enzyme catalyzes the reduction of two farnesyl pyrophosphate into squalene in ergosterol biosynthesis pathway. Ergosterol is the well-known target for many antifungal agents and depletion in ergosterol content affects major cellular processes and membrane properties74. Moreover, farnesol (200 μM) is the quorum sensing signaling molecule produced by Candida spp., which down regulates the expression of ERG9, ERG20 and ERG1175. Similarly, bafilomycin C1 compound, isolated from Streptomyces albolongus, down regulates the expression of Erg9 and other genes involved in ergosterol biosynthetic pathway76. In the present study, oleic acid down regulated the expression of Erg9p by 1.5 fold and this result was further substantiated by in vitro quantification of ergosterol and qPCR analysis.
In C. albicans, secreted lipase gene family contains 10 members (LIP1-LIP10)77. Hydrolytic enzymes such as SAPs, phospholipases and lipases are considered as important virulence factors of C. albicans which are responsible for the cell membrane damage in host and thereby promoting adhesion, invasion and colonization78. Down regulation of Lip1p (1.5 fold) by oleic acid treatment was further validated by in vitro experiment for lipase production. Likewise, the farnesol related compound, cis-2-dodecenoic acid decreased the expression of LIP1 (36.2%) of C. albicans79.
Peroxiredoxin (Tsa1B) is a thiol-specific peroxidase that catalyzes the reduction of hydrogen peroxide to water and organic hydroperoxides to alcohol. Tsa1p is differentially localized in C. albicans, wherein cell wall of hyphae and nucleus of yeast and Tsa1p are essential for the proper organization of cell wall and provide resistance to oxidative stress80,81. In C. albicans, two cytosolic Mn Superoxide Dismutases (SOD1 and SOD3) and one mitochondrial CuZn SOD (SOD2) prevent the cells from ROS43. H2O2 sensitivity assay results confirmed that increasing concentration of oleic acid (40, 80 and 160 μg mL−1) treated Candida cells are more sensitive to H2O2. Inhibitory effect of oleic acid on yeast-to-hyphal transition and oxidative stress tolerance might be the reason for the down regulation of Tsa1b (1.5 fold). This result goes in parallel with our previous report wherein higher concentration of myristic acid (125 and 250 μg mL−1) sensitizes the C. albicans to H2O243.
In the present study, up regulation (3.0 fold) of Rex2p was observed in C. albicans upon oleic acid treatment. Rex2p (orf19.1466) was found to be involved in transcription (3’-5’-exoribonuclease activity) nucleic acid binding and ribosome biogenesis in eukaryotes82. Up regulation of Rex2p suggests that oleic acid did not affect the transcription and ribosome binding processes. The next important protein up regulated (1.9 fold) in the present study was serine hydroxymethyltransferase (Shm2) which catalyzes the interconversion of serine and glycine. Glycine is a small amino acid which plays a crucial role in one carbon metabolism thus promoting cell division83,84. In the present study, oleic acid did not affect amino acid biosynthesis of C. albicans and that might be the reason for the up regulation of Shm2p. Another up regulated protein Hexokinase-2 (1.9 fold) is a well-known phosphorylating enzyme that initiates glycolysis metabolic pathway by converting glucose to glucose 6-phosphate. Upregulation of Rex2p, Shmp and Hxk2p further substantiates the non-antimicrobial effect of oleic acid against C. albicans which has also been corroborated through antimicrobial assay and XTT assay.
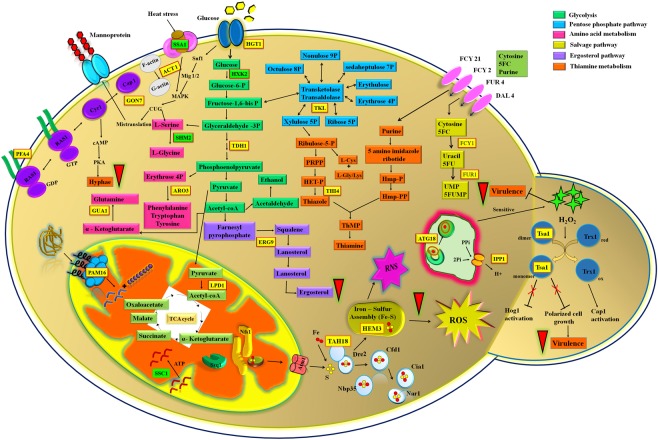
Heat shock proteins (Hsps) exist in most of the organisms and protect the organism from biotic and abiotic stresses85. In C. albicans, Hsps are found to be involved in MAPK, Ras1-cAMP-PKA, calcium-calcineurin, and cell cycle control signaling pathways. Recent studies reported that Hsps (Hsp70 and Hsp90) inhibit normal growth of C. albicans and thereby confer drug resistance to the pathogen86. Moreover, Sapiro et al.87 reported that pharmacological inhibition of Hsp90 by geldanamycin induced the filamentous growth of C. albicans in liquid rich medium at 30 °C. Consequently, in the present study, oleic acid exhibits significant inhibition on hyphal growth of C. albicans and that might be the reason for the up regulation of heat shock protein Ssa1 or Hsp70 (1.6 fold). Yet another Hsp up regulated in the present study was mitochondrial heat shock protein Ssc1, which is involved in protein transport in mitochondria. Besides, Ssc1p expression was up regulated in electron transport chain complex I mutants (nuo1Δ and nuo2Δ) which responds to amino acid starvation88. Hence, Hsps and Hsps associated pathways are seen as novel antifungal targets against candidiasis treatment. In the present study, up regulation of Ssc1p (1.5 fold) was observed which suggests that oleic acid did not affect the mitochondrial protein import, protein folding and energy metabolism. Schematic representation of differentially regulated proteins and pathways targeted by oleic acid in C. albicans is depicted in Fig. 8.
Figure 8.
Schematic representation of the proteins and pathways of C. albicans embattled by oleic acid. Up regulated and down regulated protein spots are highlighted in green and yellow boxes, respectively.
On the whole, the present study reports the inhibitory efficacy of oleic acid on C. albicans virulence including biofilm formation. In vitro virulence assays revealed the inhibitory effect of oleic acid on Candida spp. biofilm development, morphological transition, secreted hydrolases production and ergosterol biosynthesis. These results were further substantiated by gene expression studies. Further, proteomic analysis of C. albicans revealed that oleic acid exerts stress conditions such as heat stress, ROS and RNS and also targets proteins involved in basic pathways such as glucose metabolism, nucleic acid, amino acid and vitamin biosynthesis. Hence, these proteins are considered as highly conserved and they are speculated to be the key players involved in anti-virulence activity of oleic acid in C. albicans. Thus, the present study suggests that oleic acid could be used as an ideal agent for the treatment of biofilm associated infection caused by Candida spp.
Methods
Ethics statement
The present study was carried out in reference to the recommendations of Ethical Guidelines for Biomedical Research on Human Subjects, issued by Indian Council of Medical Research. The protocol was approved by the Institutional Ethics Committee, Alagappa University (Ref No: IEC/AU/2014/2). All participants gave written informed approval in accordance with the Declaration of Helsinki.
Fungal strains and growth conditions
In this study, C. albicans ATCC 90028, C. glabrata MTCC3019, C. tropicalis MTCC 184, C. albicans clinical isolates CA1 (MF423465), CA2, CA3 (MF423466), CA4 (MF423467) and C. tropicalis clinical isolates CT1 (MF423462), CT2 (MF423463), CT3 (MF423464) were used. The clinical isolates were collected from patients with Candida infection, identified at the species level by ITS sequencing. All the strains were maintained on Sabouraud dextrose agar (SDA) (Himedia, India) plates at 4 °C. To determine the effect of oleic acid on Candida spp. growth and biofilm, YEPD (1% Yeast extract, 2% glucose, and 2% Peptone) and spider broth (1% Mannitol, 0.2% K2HPO4, and 1% Nutrient broth) were used, respectively. For filamentation assay, spider agar was used. All the culture media components used in the present study were procured from Himedia, India and all the reagents used for the proteomics study were procured from Sigma, USA.
Antibiofilm assay and microscopic observation of biofilms
In order to check the antibiofilm efficacy of oleic acid on Candida spp., biofilm susceptibility assay was performed using the method as previously described by our group17. In brief, Candida spp. biofilm was grown in spider broth in the absence and presence of oleic acid (at different concentrations 5, 10, 20, 40, 80, 160 and 320 μg mL−1) in 24 well MTP at 37 °C for 24 h. After incubation, loosely adherent planktonic cells were washed with distilled water and the sessile biofilm cells were stained with 0.4% crystal violet for 5 min followed by washing off the excess stain with distilled water. Finally, 1 mL of glacial acetic acid was used to dissolve the crystal violet present in biofilm cells and absorbance was measured at 570 nm. Biofilms formed on glass surfaces in the presence and absence of oleic acid were visualized under light and confocal laser scanning microscopes. Detailed procedures are provided in Supplementary Data.
Antimicrobial assay and XTT assay
To determine the antifungal activity of oleic acid at BIC against Candida spp. growth, absorbance at 600 nm was measured using multifunctional spectrometer. In addition, XTT reduction assay was performed to assess the effect of oleic acid on Candida spp. cell viability15. Detailed procedures are provided in Supplementary Data.
Filamentation assay
The effect of oleic acid on Candida spp. filamentation was assessed using spider agar medium containing 1% fetal bovine serum (FBS) and the filamentous growth of Candida spp. were photographed. Detailed procedures are provided in Supplementary Data.
EPS extraction and quantification
EPS was extracted from oleic acid treated and untreated Candida strains by the method followed by our previous study15 with slight changes. Total carbohydrates, lipids, protein and eDNA level in EPS were quantified using phenol sulfuric acid method (optical density (OD) at 490 nm), phospho-vanillin method (OD at 545 nm), Bradford method (OD at 595 nm) and nano spectrophotometer (OD at 260/280 ratio), respectively. Detailed procedures are provided in Supplementary Data.
Quantification of SAPs and lipases
SAPs and lipase production was qualitatively measured using bovine serum albumin (BSA) agar and tributyrin agar, respectively. White opaque zone around the colonies represent SAPs production and zone of clearance around the colonies indicate lipase production. Detailed procedures are provided in Supplementary Data.
Ergosterol extraction
Changes in the ergosterol content in the absence and presence of oleic acid was measured spectrometrically3. Ergosterol was extracted from the control and oleic acid treated Candida spp. and scanned spectrophotometrically between 200 and 300 nm. Detailed procedures are provided in Supplementary Data.
Adhesion assay
Alamar blue assay was performed to assess the adhesion ability of C. albicans on polystyrene surfaces in the absence and presence of oleic acid at different concentration (10, 20, 40, 80 and 160 μg mL−1). The control and oleic acid treated cells were incubated with alamar blue solution and the fluorescent intensity was measured at 590 nm emission and 560 nm excitation wavelengths89. Detailed procedures are provided in Supplementary Data.
H2O2 sensitivity assay
Overnight grown C. albicans culture in the presence and absence of oleic acid (80 and 160 μg mL−1) OD600 nm was adjusted to 0.3 and swabbed on YEPD agar plates. To assess the H2O2 sensitivity, sterile filter paper disks with 10 mm diameter (Himedia, India) were placed on YEPD agar plates, loaded with 15 μL of 30% H2O2 and incubated at 37 °C for 16 h. After incubation, zone of clearance was measured and the plates were documented43.
Real Time PCR
Total RNA was isolated from control and oleic acid treated (at BIC) cultures using hot phenol extraction method15 and converted into cDNA using High capacity cDNA Reverse Transcription kit (Applied Biosystems, USA). Candidate virulence genes (als1, als3, cdr1, mdr1, erg11, flu1,nrg1, sap1, sap2, sap4, tup1, hwp1, eap1, efg1, cst20, ras1, ume6, hst7 and cph1, chs3, cht4,) were selected for real time PCR experiment (7500 Sequence Detection System, Applied Biosystems Inc. Foster, CA, USA). The primers were combined individually with SYBR Green kit (Applied Biosystems, USA) at a predefined ratio. The expression pattern of candidate genes were normalized against ITS gene (~540 bp) expression (housekeeping gene) and quantified using the ΔΔCT method. Detailed procedures are provided in Supplementary Data.
Intracellular protein extraction
For intracellular protein extraction, C. albicans was grown in the absence and presence of oleic acid (80 μg mL−1) in YEPD supplemented with FBS and incubated at 37 °C in a shaking condition for 16 h (until the cells reach mid log phase). Then, the cells were collected, by centrifugation (8000 rpm, 20 min at 4 °C) and washed twice with PBS (pH – 7.4) and sonicated in 20 mM Tris-HCl (pH – 8.0) containing 1% protease inhibitor cocktail, 100 mM PMSF and 1 mM of 0.5 M EDTA. After sonication, cell suspension was centrifuged (13000 rpm, 30 min at 4 °C) and the supernatant was collected. To purify the cellular proteins, equal volume of phenol was added to the supernatant, and incubated at 70 °C for 10 min and cooled at 4 °C for 10 min. Then, equal volume of milli Q water was added to the phenol/protein extract mixture, vortexed and incubated as mentioned in the previous step. Subsequently, the tubes were centrifuged (8,000 rpm, 10 min at 4 °C) for phase separation. The aqueous phase was discarded; protein present in the phenol phase was precipitated with double the volume of ice cold acetone. Then, the precipitated proteins were collected by centrifugation (12,000 rpm, 15 min at 4 °C) and washed thrice with acetone to remove the residual phenol. After thrice washing with acetone, the pellets air dried. Then, the dried protein pellets were dissolved in sample buffer containing 7 M Urea, 2 M thiourea and 4% CHAPS11.
Two dimensional gel electrophoresis
Two dimensional gel electrophoresis (2DGE) was performed using the method described in our previous report11. Briefly, first dimension/isoelectric focusing (IEF) was performed in IPGphor 3 system using immobiline DryStrip gel strips (18 cm, non- linear, pH 4–7). Protein samples (each 1000 μg from control and oleic acid treated) were mixed with rehydration buffer containing 7 M urea, 2 M thiourea, 2% (CHAPS), 12.5 mg mL−1 destreak reagent and 0.5% IPG buffer (pH 4–7) to a final volume of 350 μL and applied to the IPG strips by in- gel rehydration for 16 h at 20 °C. After rehydration, IPG strips containing protein samples were subjected to isoelectric focusing at 20 °C under mineral oil using following conditions: 2 h at 100 V; 3 h at 500 V (gradient); 3 h at 500 V; 2.5 h at 5000 V (gradient); 2 h at 5000 V (gradient); 3 h at 8000 V (gradient); 2 h at 8000 V and final focussing was done for 2 h at 10000 V. The current was set to 75 μA per IPG strip. After IEF, the IPG strips were incubated in equilibration buffer I [6 M urea, 30% (w/v) glycerol, 2% (w/v) sodium dodecyl sulphate (SDS) and 1% (w/v) DTT in 50 mM Tris-HCl buffer, pH 8.8] followed by equilibration buffer II [6 M urea, 30% (w/v) glycerol, 2% (w/v) SDS and 2.5% (w/v) iodacetamide (IAA) in 50 mM Tris-HCl buffer, pH 8.8] for 15 min each. After two equilibration steps, for second dimension, IPG strips were placed on 22 cm × 22 cm × 1 mm 10–15% gradient sodium dodecyl sulphate- polyacrylamide gels (SDS-PAGE) and overlaid with 0.3% agarose. Electrophoresis was performed at 100 V for 1 h and 150 V for 8 h in Ettan DALT six apparatus (GE Healthcare, USA). After electrophoresis, gels were immersed in fixative for 3 h and washed thrice with milli Q water for 20 min each. Protein spots were stained using CBB G-250 staining solution for 12 h on rotary agitator. After staining, gels were destained with milli Q water for 4 h to reduce the background11.
Protein spot excision and trypsin digestion
Differentially regulated protein spots with more than 1.5 fold changes in the intensity (both up and down regulated) were selected and excised from the gels for protein identification. Briefly, prior to destaining, gel pieces were washed with milli Q water. Then, the gel pieces were completely destained by washing with destaining solution containing 50% acetonitrile containing 25 mM ammonium bicarbonate (NH4HCO3). Destained gel pieces were completely dehydrated in 100% acetonitrile (ACN) for 10 min and dried under vacuum for 30 min. Dried gel pieces were incubated in reduction solution (10 mM DTT and 25 mM NH4HCO3) followed by alkylation solution (25 mM NH4HCO3 and 55 mM IAA) for 30 min each. After reduction and alkylation step, the gel pieces were dehydrated with 100 μL of acetonitrile and dried under vacuum for 30 min. Then, the gel pieces were rehydrated/trypsinized with 5 μL of digestion buffer (10 mM NH4HCO3 in 10% ACN) containing 400 ng of trypsin (Sigma Aldrich) on ice for 30 min. Then, the rehydrated gel pieces were covered with 25 μL of overlay buffer (40 mM NH4HCO3 in 10% ACN) and incubated at 37 °C for 16 h. After incubation, peptides were extracted twice with 25 μl of 0.1% trifluoroacetic acid (TFA) in 60% ACN by sonication (10 min) followed by 20 μL of 100% ACN. Extracted peptides were dried under vacuum for 90 min and stored at 4 °C11. Before MALDI-TOF/TOF analysis, peptides were dissolved in peptide resuspension solution containing 0.1% TFA in 5% ACN. Then, the salts present in the peptides were removed using C18 zip tips (Merck Millipore), the desalted peptides were dried and stored at 4 °C.
MALDI- TOF/TOF analysis
Differentially regulated proteins upon oleic acid treatment were analyzed using MALDI- TOF/TOF analysis13. Briefly, equal volume (1 μL) of peptides were mixed with 1 μL of matrix solution [alpha-cyano-4-hydroxy cinnamic acid matrix (10 mg mL−1), acetonitrile (50%) and trifluroacetic acid (0.1%)] and spotted on MALDI target plate. The MALDI- TOF/TOF mass spectrometer (AXIMA Performance, SHIMADZU BIOTECH) with laser wavelength of 337 nm was calibrated using TOF- Mix™ (LaserBio Labs, France). Peptide mass spectra were acquired from a MALDI- TOF/TOF mass spectrometer in positive reflector mode and analysed by Shimadzu launch pad- MALDI MS software (Version- 2.9.3.20110624, Kratos Analytical Ltd, U.K.). For single spot, 45 subspectra resulted after 50 laser shots at randomized positions with total of 2250 laser shots. The laser intensity for MS spectrum acquisition was 4500. Mono isotopic peak list (m/z range of 700–4000 Da with S/N ratio over 10) generated by MALDI MS software was analyzed using online software tool MS-Fit from Protein Prospector (v 5.24.0). (http://prospector.ucsf.edu/prospector/cgi-bin/msform.cgi?form=msfitstandard) with the standard protein identification parameters. Briefly, a mass tolerance of 1.50 Da per peptide, maximum of 2 missed cleavages per peptide, carbamidomethylation of cysteine was given as fixed modification whereas N-terminal acetylation and phosphorylation (S, T, Y) and methionine oxidation, as variable modifications.
Statistical analysis
All the experiments were performed at least twice in independent experiments in triplicates to confirm reproducibility and the data were presented as mean ± standard deviation. For all experiments, statistical differences between control and treated samples were analyzed with one way ANOVA followed by Dunnett’s test with significant p-value <0.05 using SPSS software version 17.0 (Chicago, IL, USA).
Supplementary information
Acknowledgements
The authors sincerely acknowledge the computational and bioinformatics facility provided by the Bioinformatics Infrastructure Facility (funded by DBT, GOI; File No. BT/BI/25/012/2012, BIF). The authors also thankfully acknowledge DST-FIST (Grant No. SR/FST/LSI-639/2015(C)), UGC-SAP (Grant No. F.5-1/2018/DRS-II (SAP-II)), DST-PURSE (Grant No. SR/PURSE Phase 2/38 (G)) and RUSA 2.0 (Grant No. F. 24-51/2014-U, Policy (TN Multi-Gen), Dept of Edn, GOI) for providing instrumentation facilities. SM thanks UGC for financial assistance in the form of a Basic Scientific Research Fellowship [Sanction No. F.25-1/2013-14 (BSR)/7-326/2011(BSR)]. S.K.P. is thankful to UGC for Mid-Career Award [F.19-225/2018(BSR)].
Author contributions
S.K.P. and S.M. conceived the experiments, S.M., K.G.P., A.P. and P.P. conducted the experiments, S.M. and S.K.P. wrote the main manuscript text and S.M. prepared the figures. All authors reviewed the manuscript.
Data availability
The raw data of MALDI-TOF/TOF was submitted in ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD017588.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Subramanian Muthamil and Krishnan Ganesh Prasath.
Supplementary information
is available for this paper at 10.1038/s41598-020-61918-y.
References
- 1.Nett J, Andes D. Candida albicans biofilm development, modeling a host–pathogen interaction. Curr. Opin. Microbiol. 2006;9:340–345. doi: 10.1016/j.mib.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Odds FC, Webster CE, Mayuranathan P, Simmons PD. Candida concentrations in the vagina and their association with signs and symptoms of vaginal candidosis. J. Med. Vet. Mycol. 1988;26:277–283. doi: 10.1080/02681218880000391. [DOI] [PubMed] [Google Scholar]
- 3.Shafreen RM, Muthamil S, Pandian SK. Inhibition of Candida albicans virulence factors by novel levofloxacin derivatives. Appl. Microbiol. Biotechnol. 2014;98:6775–6785. doi: 10.1007/s00253-014-5719-2. [DOI] [PubMed] [Google Scholar]
- 4.Rathna J, Bakkiyaraj D, Pandian SK. Anti-biofilm mechanisms of 3, 5-di-tert-butylphenol against clinically relevant fungal pathogens. Biofouling. 2016;32:979–993. doi: 10.1080/08927014.2016.1216103. [DOI] [PubMed] [Google Scholar]
- 5.Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowan MM. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 1999;12:564–582. doi: 10.1128/CMR.12.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Upadhyay Abhinav, Upadhyaya Indu, Kollanoor-Johny Anup, Venkitanarayanan Kumar. Combating Pathogenic Microorganisms Using Plant-Derived Antimicrobials: A Minireview of the Mechanistic Basis. BioMed Research International. 2014;2014:1–18. doi: 10.1155/2014/761741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashwinkumar Subramenium Ganapathy, Viszwapriya Dharmaprakash, Iyer Prasanth Mani, Balamurugan Krishnaswamy, Karutha Pandian Shunmugiah. covR Mediated Antibiofilm Activity of 3-Furancarboxaldehyde Increases the Virulence of Group A Streptococcus. PLOS ONE. 2015;10(5):e0127210. doi: 10.1371/journal.pone.0127210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subramenium Ganapathy Ashwinkumar, Vijayakumar Karuppiah, Pandian Shunmugiah Karutha. Limonene inhibits streptococcal biofilm formation by targeting surface-associated virulence factors. Journal of Medical Microbiology. 2015;64(8):879–890. doi: 10.1099/jmm.0.000105. [DOI] [PubMed] [Google Scholar]
- 10.Sethupathy S, Nithya C, Pandian SK. 2-Furaldehyde diethyl acetal from tender coconut water (Cocos nucifera) attenuates biofilm formation and quorum sensing-mediated virulence of Chromobacterium violaceum and Pseudomonas aeruginosa. Biofouling. 2015;31:721–733. doi: 10.1080/08927014.2015.1102897. [DOI] [PubMed] [Google Scholar]
- 11.Sethupathy S, et al. Proteomic analysis reveals modulation of iron homeostasis and oxidative stress response in Pseudomonas aeruginosa PAO1 by curcumin inhibiting quorum sensing regulated virulence factors and biofilm production. J. Proteomics. 2016;145:112–126. doi: 10.1016/j.jprot.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 12.Salini R, Pandian SK. Interference of quorum sensing in urinary pathogen Serratia marcescens by Anethum graveolens. Pathog. Dis. 2015;73:ftv038. doi: 10.1093/femspd/ftv038. [DOI] [PubMed] [Google Scholar]
- 13.Sethupathy, S. et al. Vanillic acid from Actinidia deliciosa impedes virulence in Serratia marcescens by affecting S-layer, flagellin and fatty acid biosynthesis proteins. Sci. Rep. 7, 16328, https://www.nature.com/articles/s41598-017-16507-x (2017). [DOI] [PMC free article] [PubMed]
- 14.Sivasankar C, Gayathri S, Bhaskar JP, Krishnan V, Pandian SK. Evaluation of selected Indian medicinal plants for antagonistic potential against Malassezia spp. and the synergistic effect of embelin in combination with ketoconazole. Microb. Pathog. 2017;110:66–72. doi: 10.1016/j.micpath.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 15.Muthamil, S., Balasubramaniam, B., Balamurugan, K. & Pandian, S. K. Synergistic effect of quinic acid derived from Syzygium cumini and undecanoic acid against Candida spp. biofilm and virulence. Front. Microbiol. 9, 10.3389/fmicb.2018.02835 (2018). [DOI] [PMC free article] [PubMed]
- 16.Agoramoorthy G, Chandrasekaran M, Venkatesalu V, Hsu MJ. Antibacterial and antifungal activities of fatty acid methyl esters of the blind-your-eye mangrove from India. Braz. J. Microbiol. 2007;38:739–742. doi: 10.1590/S1517-83822007000400028. [DOI] [Google Scholar]
- 17.Desbois AP, Lawlor KC. Antibacterial activity of long-chain polyunsaturated fatty acids against Propionibacterium acnes and Staphylococcus aureus. Mar. Drugs. 2013;11:4544–4557. doi: 10.3390/md11114544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereira de Mello T, de Souza Ramos L, Adriana Braga-Silva L, Helena Branquinha M, Luis Souza dos Santos A. Fungal biofilm–a real obstacle against an efficient therapy: lessons from Candida. Curr. Top. Med. Chem. 2017;17:1987–2004. doi: 10.2174/1568026617666170105145227. [DOI] [PubMed] [Google Scholar]
- 19.McCarty TP, Pappas PG. Invasive candidiasis. Infect. Dis. Clin. 2016;30:103–124. doi: 10.1016/j.idc.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Terra L, et al. Mycoses and Antifungals: reviewing the basis of a current problem that still is a biotechnological target for marine products. Front. Mar. Sci. 2014;1:12. doi: 10.3389/fmars.2014.00012. [DOI] [Google Scholar]
- 21.Sivasankar C, et al. A combination of ellagic acid and tetracycline inhibits biofilm formation and the associated virulence of Propionibacterium acnes in vitro and in vivo. Biofouling. 2016;32:397–410. doi: 10.1080/08927014.2016.1148141. [DOI] [PubMed] [Google Scholar]
- 22.Slobodnikova L, Fialova S, Rendekova K, Kovac J, Mucaji P. Antibiofilm Activity of Plant Polyphenols. Molecules. 2016;21:1717. doi: 10.3390/molecules21121717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banu SF, et al. Antivirulent properties of underexplored Cinnamomum tamala essential oil and its synergistic effects with DNase against Pseudomonas aeruginosa biofilms-an in vitro study. Front. Microbiol. 2017;8:1144. doi: 10.3389/fmicb.2017.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandit S, Cai JN, Song KY, Jeon JG. Identification of anti‐biofilm components in Withania somnifera and their effect on virulence of Streptococcus mutans biofilms. J. Appl. Microbiol. 2015;119:571–581. doi: 10.1111/jam.12851. [DOI] [PubMed] [Google Scholar]
- 25.Shebl Rania Ibrahim, Farouk Faten, Azzazy Hassan Mohamed El-Said. Effect of Surface Charge and Hydrophobicity Modulation on the Antibacterial and Antibiofilm Potential of Magnetic Iron Nanoparticles. Journal of Nanomaterials. 2017;2017:1–15. doi: 10.1155/2017/3528295. [DOI] [Google Scholar]
- 26.Chen CH, et al. An innate bactericidal oleic acid effective against skin infection of methicillin-resistant Staphylococcus aureus: a therapy concordant with evolutionary medicine. J. Microbiol. Biotechnol. 2011;21:391–399. doi: 10.4014/jmb.1011.11014. [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen TB, Givskov M. Quorum-sensing inhibitors as antipathogenic drugs. Int. J. Med. Microbiol. 2006;296:149–161. doi: 10.1016/j.ijmm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Era M, et al. Antifungal activity of fatty acid salts against Penicillium pinophilum. Jpn. J. Food Eng. 2015;16:99–108. doi: 10.11301/jsfe.16.99. [DOI] [Google Scholar]
- 29.Kaur N, Garg R. Optimization and evaluation of oleic acid based unsaturated fatty acid liposomes gel. J. Bioequiv. Availab. 2017;9:424–429. doi: 10.4172/jbb.1000336. [DOI] [Google Scholar]
- 30.Goffena J, Toenjes KA, Butler DK. Inhibition of yeast‐to‐filamentous growth transitions in Candida albicans by a small molecule inducer of mammalian apoptosis. Yeast. 2018;35:291–298. doi: 10.1002/yea.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muthamil S, Pandian SK. Inhibitory effect of Murraya koenigii against Candida albicans virulence and biofilm development. Biologia. 2016;71:256–264. doi: 10.1515/biolog-2016-0044. [DOI] [Google Scholar]
- 32.Manoharan RK, Lee JH, Lee J. Efficacy of 7‐benzyloxyindole and other halogenated indoles to inhibit Candida albicans biofilm and hyphal formation. Microb. Biotechnol. 2018;11:1060–1069. doi: 10.1111/1751-7915.13268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padmavathi AR, Abinaya B, Pandian SK. Phenol, 2, 4-bis (1, 1-dimethylethyl) of marine bacterial origin inhibits quorum sensing mediated biofilm formation in the uropathogen Serratia marcescens. Biofouling. 2014;30:1111–1122. doi: 10.1080/08927014.2014.972386. [DOI] [PubMed] [Google Scholar]
- 34.Nithyanand P, Beema Shafreen RM, Muthamil S, Pandian SK. Usnic acid inhibits biofilm formation and virulent morphological traits of Candida albicans. Microbiol. Res. 2015;179:20–28. doi: 10.1016/j.micres.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 35.Subramenium GA, Swetha TK, Iyer PM, Balamurugan K, Pandian SK. 5- hydroxymethyl-2-uraldehyde from marine bacterium Bacillus subtilis inhibits biofilm and virulence of Candida albicans. Microbiol. Res. 2018;207:19–32. doi: 10.1016/j.micres.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 36.Kantarcioglu AS, Yucel A. Phospholipase and protease activities in clinical Candida isolates with reference to the sources of strains. Mycoses. 2002;45:160–165. doi: 10.1046/j.1439-0507.2002.00727.x. [DOI] [PubMed] [Google Scholar]
- 37.Mohandas, V. & Ballal, M. Distribution of Candida species in different clinical samples and their virulence: biofilm formation, proteinase and phospholipase production: a study on hospitalized patients in southern India. J. Glob. Infect. Dis. 3, 4–8, http://www.jgid.org/text.asp?2011/3/1/4/77288 (2011). [DOI] [PMC free article] [PubMed]
- 38.Hamid S, Zainab S, Faryal R, Ali N, Sharafat I. Inhibition of secreted aspartyl proteinase activity in biofilms of Candida species by mycogenic silver nanoparticles. Artif. Cell. Nanomed, B. 2018;46:551–557. doi: 10.1080/21691401.2017.1328688. [DOI] [PubMed] [Google Scholar]
- 39.Xu Y, et al. Requirement for Ergosterol in Berberine tolerance underlies synergism of Fluconazole and Berberine against fluconazole-resistant Candida albicans isolates. Front. Cell. Infect. Microbiol. 2017;7:491. doi: 10.3389/fcimb.2017.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuza-Alves DL, Silva-Rocha WP, Chaves GM. An update on Candida tropicalis based on basic and clinical approaches. Front. Microbiol. 2017;8:1927. doi: 10.3389/fmicb.2017.01927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masood MM, et al. 2, 3-Triazole–quinazolin-4 (3 H)-one conjugates: evolution of ergosterol inhibitor as anticandidal agent. RSC Adv. 2018;8:39611–39625. doi: 10.1039/C8RA08426B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu CC, Lai WL, Chuang KC, Lee MH, Tsai YC. The inhibitory activity of linalool against the filamentous growth and biofilm formation in Candida albicans. Med. Mycol. 2013;51:473–482. doi: 10.3109/13693786.2012.743051. [DOI] [PubMed] [Google Scholar]
- 43.Prasath KG, Sethupathy S, Pandian SK. Proteomic analysis uncovers the modulation of ergosterol, sphingolipid and oxidative stress pathway by myristic acid impeding biofilm and virulence in Candida albicans. J. Proteomics. 2019;208:103503. doi: 10.1016/j.jprot.2019.103503. [DOI] [PubMed] [Google Scholar]
- 44.Vylkova S, et al. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio. 2011;2:e00055–11. doi: 10.1128/mBio.00055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherrington SL, et al. Adaptation of Candida albicans to environmental pH induces cell wall remodelling and enhances innate immune recognition. PLoS Pathog. 2017;13:e1006403. doi: 10.1371/journal.ppat.1006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukherjee PK, et al. Alcohol dehydrogenase restricts the ability of the pathogen Candida albicans to form a biofilm on catheter surfaces through an ethanol-based mechanism. Infect. Immun. 2006;74:3804–3816. doi: 10.1128/IAI.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siikala E, et al. ADH1 expression inversely correlates with CDR1 and CDR2 in Candida albicans from chronic oral candidosis in APECED (APS-I) patients. FEMS Yeast Res. 2011;11:494–498. doi: 10.1111/j.1567-1364.2011.00739.x. [DOI] [PubMed] [Google Scholar]
- 48.Chauhan NM, Raut JS, Karuppayil SM. Acetaldehyde inhibits the yeast-to-hypha conversion and biofilm formation in Candida albicans. Mycoscience. 2011;52:356–360. doi: 10.1007/S10267-011-0110-Y. [DOI] [Google Scholar]
- 49.Nieminen MT, et al. A novel antifungal is active against Candida albicans biofilms and inhibits mutagenic acetaldehyde production in vitro. PLoS One. 2014;9:e97864. doi: 10.1371/journal.pone.0097864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandra J, et al. In vitro and in vivo activity of a novel catheter lock solution against bacterial and fungal biofilms. Antimicrob. Agents Chemother. 2018;62:e00722–18. doi: 10.1128/AAC.00722-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Almeida RS, et al. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 2008;4:e1000217. doi: 10.1371/journal.ppat.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fourie, R., Kuloyo, O. O, Mochochoko, B. M., Albertyn, J. & Pohl, C. H. Iron at the Centre of Candida albicans Interactions. Front. Cell. Infect. Microbiol. 8, 10.3389/fcimb.2018.00185 (2018). [DOI] [PMC free article] [PubMed]
- 53.Zhou S, et al. Heme-biosynthetic porphobilinogen deaminase protects Aspergillus nidulans from nitrosative stress. Appl. Environ. Microbiol. 2012;78:103–109. doi: 10.1128/AEM.06195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maidan MM, Thevelein JM, Van Dijck P. Carbon source induced yeast-to-hypha transition in Candida albicans is dependent on the presence of amino acids and on the G-protein-coupled receptor Gpr1. Biochem. Soc. T. 2005;33:291–293. doi: 10.1042/BST0330291. [DOI] [PubMed] [Google Scholar]
- 55.Pemmaraju SC, Pruthi PA, Prasad R, Pruthi V. Modulation of Candida albicans biofilm by different carbon sources. Mycopathologia. 2016;181:341–352. doi: 10.1007/s11046-016-9992-8. [DOI] [PubMed] [Google Scholar]
- 56.Rodriguez-Suarez R, et al. Mechanism-of-action determination of GMP synthase inhibitors and target validation in Candida albicans and Aspergillus fumigatus. Chem. Biol. 2007;14:1163–1175. doi: 10.1016/j.chembiol.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 57.Jiang L, et al. Functional characterization and virulence study of ADE8 and GUA1 genes involved in the de novo purine biosynthesis in Candida albicans. FEMS Yeast Res. 2010;10:199–208. doi: 10.1111/j.1567-1364.2009.00600.x. [DOI] [PubMed] [Google Scholar]
- 58.Chitty JL, et al. GMP synthase is required for virulence factor production and infection by Cryptococcus neoformans. J. Biol. Chem. 2017;292:3049–3059. doi: 10.1074/jbc.M116.767533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin Z, et al. Proteomic response to amino acid starvation in Candida albicans and Saccharomyces cerevisiae. Proteomics. 2004;4:2425–2436. doi: 10.1002/pmic.200300760. [DOI] [PubMed] [Google Scholar]
- 60.Martinez P, Ljungdahl PO. An ER packaging chaperone determines the amino acid uptake capacity and virulence of Candida albicans. Mol. Microbiol. 2004;51:371–384. doi: 10.1046/j.1365-2958.2003.03845.x. [DOI] [PubMed] [Google Scholar]
- 61.Garbe E, Vylkova S. Role of Amino Acid Metabolism in the Virulence of Human Pathogenic Fungi. Curr. Clin. Micro. Rpt. 2019;6:108–119. doi: 10.1007/s40588-019-00124-5. [DOI] [Google Scholar]
- 62.Kamthan M, Mukhopadhyay G, Chakraborty N, Chakraborty S, Datta A. Quantitative proteomics and metabolomics approaches to demonstrate N-acetyl-D-glucosamine inducible amino acid deprivation response as morphological switch in Candida albicans. Fungal Genet. Biol. 2012;49:369–378. doi: 10.1016/j.fgb.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 63.Herrero AB, Uccelletti D, Hirschberg CB, Dominguez A, Abeijon C. The Golgi GDPase of the fungal pathogen Candida albicans affects morphogenesis, glycosylation, and cell wall properties. Eukaryot. Cell. 2002;1:420–431. doi: 10.1128/EC.1.3.420-431.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cantero PD, Ernst JF. Damage to the glycoshield activates PMT‐directed O‐mannosylation via the Msb2–Cek1 pathway in Candida albicans. Mol. Microbiol. 2011;80:715–725. doi: 10.1111/j.1365-2958.2011.07604.x. [DOI] [PubMed] [Google Scholar]
- 65.Gil-Bona A, et al. Candida albicans cell shaving uncovers new proteins involved in cell wall integrity, yeast to hypha transition, stress response and host–pathogen interaction. J. Proteomics. 2015;127:340–351. doi: 10.1016/j.jprot.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bramono, K., Yamazaki, M., Tsuboi, R. & Ogawa, H. Comparison of proteinase, lipase and alpha-glucosidase activities from the clinical isolates of Candida species. Jpn. J. Infect. Dis. 59, 73–76, http://www.nih.go.jp/JJID/59/73.html (2006). [PubMed]
- 67.Kim J, Bao TH, Shin YK, Kim KY. Antifungal activity of magnoflorine against Candida strains. World J. Microbiol. Biot. 2018;34:167. doi: 10.1007/s11274-018-2549-x. [DOI] [PubMed] [Google Scholar]
- 68.Yin Z, et al. A proteomic analysis of the salt, cadmium and peroxide stress responses in Candida albicans and the role of the Hog1 stress‐activated MAPK in regulating the stress‐induced proteome. Proteomics. 2009;9:4686–4703. doi: 10.1002/pmic.200800958. [DOI] [PubMed] [Google Scholar]
- 69.Meir Z, Osherov N. Vitamin biosynthesis as an antifungal target. J. Fungi. 2018;4:72. doi: 10.3390/jof4020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siemieniuk M, Czyzewska U, Strumilo S, Tylicki A. Thiamine antivitamins–an opportunity of therapy of fungal infections caused by Malassezia pachydermatis and Candida albicans. Mycoses. 2016;59:108–116. doi: 10.1111/myc.12441. [DOI] [PubMed] [Google Scholar]
- 71.Chen C, Pande K, French SD, Tuch BB, Noble SM. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe. 2011;10:118–135. doi: 10.1016/j.chom.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ror S, Panwar SL. Sef1-regulated iron regulon responds to mitochondria-dependent iron-sulfur cluster biosynthesis in Candida albicans. Front. Microbio. 2019;10:1528. doi: 10.3389/fmicb.2019.01528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nett JE, Lepak AJ, Marchillo K, Andes DR. Time course global gene expression analysis of an in vivo Candida biofilm. J. Infect. Dis. 2009;200:307–313. doi: 10.1086/599838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lv QZ, Yan L, Jiang YY. The synthesis, regulation, and functions of sterols in Candida albicans: well-known but still lots to learn. Virulence. 2016;7:649–659. doi: 10.1080/21505594.2016.1188236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dizova S, Cernakova L, Bujdakova H. The impact of farnesol in combination with fluconazole on Candida albicans biofilm: regulation of ERG20, ERG9, and ERG11 genes. Folia Microbiol. 2018;1:1–9. doi: 10.1007/s12223-017-0574-z. [DOI] [PubMed] [Google Scholar]
- 76.Su H, et al. Bafilomycin C1 exert antifungal effect through disturbing sterol biosynthesis in Candida albicans. J. Antibiot. 2018;71:467. doi: 10.1038/s41429-017-0009-8. [DOI] [PubMed] [Google Scholar]
- 77.Stehr F, et al. Expression analysis of the Candida albicans lipase gene family during experimental infections and in patient samples. FEMS Yeast Res. 2004;4:401–408. doi: 10.1016/S1567-1356(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 78.Monroy-Pérez Eric, Paniagua-Contreras Gloria Luz, Rodríguez-Purata Pamela, Vaca-Paniagua Felipe, Vázquez-Villaseñor Marco, Díaz-Velásquez Clara, Uribe-García Alina, Vaca Sergio. High Virulence and Antifungal Resistance in Clinical Strains ofCandida albicans. Canadian Journal of Infectious Diseases and Medical Microbiology. 2016;2016:1–7. doi: 10.1155/2016/5930489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liang YD, et al. Protective effects of cis-2-dodecenoic acid in an experimental mouse model of vaginal candidiasis. Biomed. Environ. Sci. 2018;31:816–828. doi: 10.3967/bes2018.109. [DOI] [PubMed] [Google Scholar]
- 80.Urban C, et al. The moonlighting protein Tsa1p is implicated in oxidative stress response and in cell wall biogenesis in Candida albicans. Mol. Microbiol. 2005;57:1318–1341. doi: 10.1111/j.1365-2958.2005.04771.x. [DOI] [PubMed] [Google Scholar]
- 81.Marin E, et al. Candida albicans shaving to profile human serum proteins on hyphal surface. Front. Microbiol. 2015;6:1343. doi: 10.3389/fmicb.2015.01343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Park H, et al. Transcriptional responses of Candida albicans to epithelial and endothelial cells. Eukaryot. Cell. 2009;8:1498–1510. doi: 10.1128/EC.00165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McNeil SD, et al. Metabolic modeling identifies key constraints on an engineered glycine betaine synthesis pathway in tobacco. Plant Physiol. 2000;124:153–162. doi: 10.1104/pp.124.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Flynn NE, Patyrak ME, Seely JB, Wu G. Glycine oxidation and conversion into amino acids in Saccharomyces cerevisiae and Candida albicans. Amino Acids. 2010;39:605–608. doi: 10.1007/s00726-010-0477-7. [DOI] [PubMed] [Google Scholar]
- 85.Tiwari Shraddha, Thakur Raman, Shankar Jata. Role of Heat-Shock Proteins in Cellular Function and in the Biology of Fungi. Biotechnology Research International. 2015;2015:1–11. doi: 10.1155/2015/132635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gong Y, Li T, Yu C, Sun S. Candida albicans heat shock proteins and Hsps-associated signaling pathways as potential antifungal targets. Front. Cell. Infect. Microbiol. 2017;7:520. doi: 10.3389/fcimb.2017.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shapiro RS, et al. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr. Biol. 2009;19:621–629. doi: 10.1016/j.cub.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.She X, et al. A mitochondrial proteomics view of complex I deficiency in Candida albicans. Mitochondrion. 2018;38:48–57. doi: 10.1016/j.mito.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 89.Selvaraj A, Jayasree T, Valliammai A, Pandian SK. Myrtenol attenuates MRSA biofilm and virulence by suppressing sarA expression dynamism. Front. Microbial. 2019;10:2027. doi: 10.3389/fmicb.2019.02027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of MALDI-TOF/TOF was submitted in ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD017588.