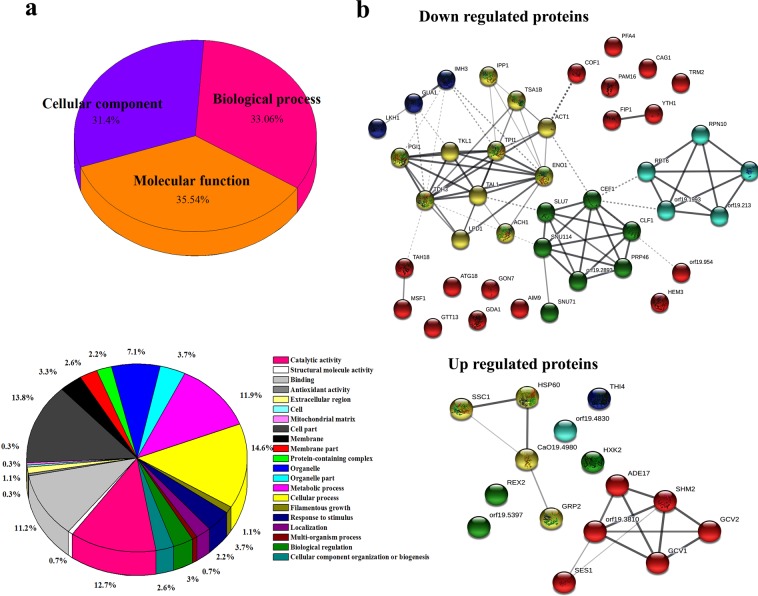
Figure 7.
Gene ontology and STRING analysis of differentially regulated proteins of C. albicans upon oleic acid treatment. (a) Gene ontology analysis of differentially regulated (both up regulated and down regulated) proteins using UniProt database. The differentially expressed proteins are mainly involved in cellular process (37%), catalytic activity (33%), metabolic process (30%), binding (29%), response to stimulus (9%), biological regulation (8%), and cellular component organization (7%) (b) Protein-protein interaction prediction map of (i) down regulated and (ii) up regulated proteins of C. albicans obtained using STRING v.11 database (confidence score 0.400). The interaction map shows the proteins involved in interrelated pathways such as proteasome, spliceosome, carbon metabolism, biosynthesis of antibiotics, biosynthesis of secondary metabolites and metabolic pathways. Filled protein nodes that signify the availability of protein 3D structural information is known or predicted.