Figure 1.
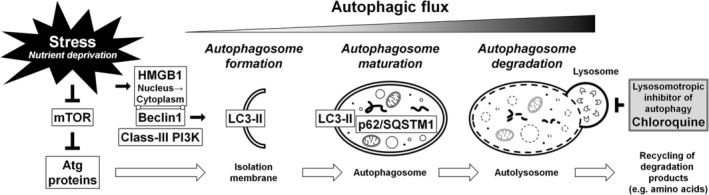
Schematic illustration of disc cellular autophagy. Under stress conditions, for example, nutrient deprivation, the mammalian target of rapamycin (mTOR), a signal integrator that detects nutrients to signal the execution of cell growth and division, is suppressed, which initiates autophagy through the activation of autophagy‐related (Atg) genes and proteins. High‐mobility group box 1 (HMGB1), which is involved in stress response, translocates from the nucleus to the cytoplasm and directly interacts with Beclin1 (Atg6 homolog). The Beclin1–class‐III phosphatidylinositol 3‐kinase (PI3K) complex initiates autophagosome formation by developing the isolation membrane. Autophagosome maturation is completed by the growth and closure of the isolation membrane, driven by the conjugation of phosphatidylethanolamine with light chain 3 (LC3) (Atg8 homolog), leading to the formation of the autophagosome‐membrane‐bound form LC3‐II. Then, p62/sequestosome 1 (p62/SQSTM1) and p62/SQSTM1‐bound polyubiquitinated proteins become incorporated into completed autophagosomes. The completed autophagosome fuses with the lysosome to form the autolysosome (which can be inhibited by chloroquine), where the enclosed cargo is degraded, and its constituents are released and recycled. Understanding of autophagy requires monitoring this dynamic, multi‐step process of autophagic flux. In our previous time‐course observational study, the graded supply of serum and nutrients decreased proliferation and metabolic activity and increased autophagy, apoptosis, and senescence in rabbit disc annulus fibrosus cells
