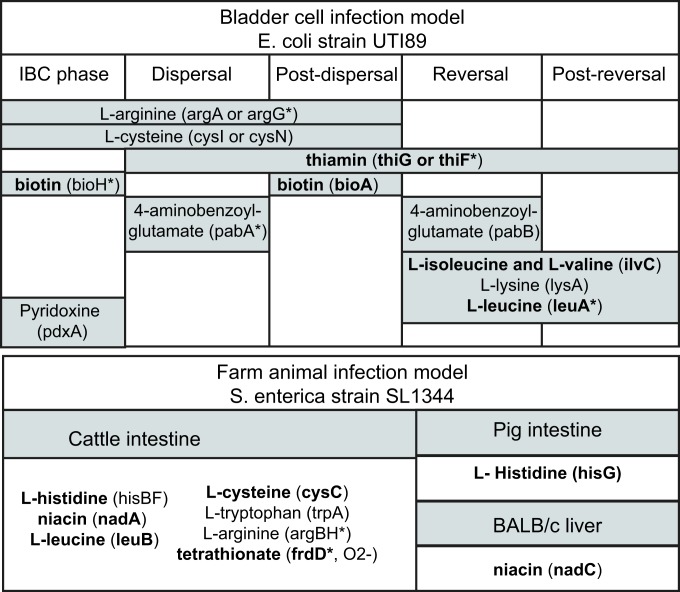
Fig. 3.
Computationally predicted CEGs that were found to result in increased fitness (more than one log2 fold change, P value < 0.05) in mutant screens. For each condition in which the mutant fitness was tested, we list out both the nutrient for which it is predicted to be auxotrophic and the gene which has been disrupted. The fitness profiles were obtained from various sources in which the TRADIS workflow was applied. The bladder cell infection model was designed as a proxy for urinary tract infection. Genes highlighted in bold are lost across natural isolates. An asterisk (*) indicates that CEG knock-out yields a detrimental effect on fitness in other conditions. See Datasets S3 and S4 for the full dataset.